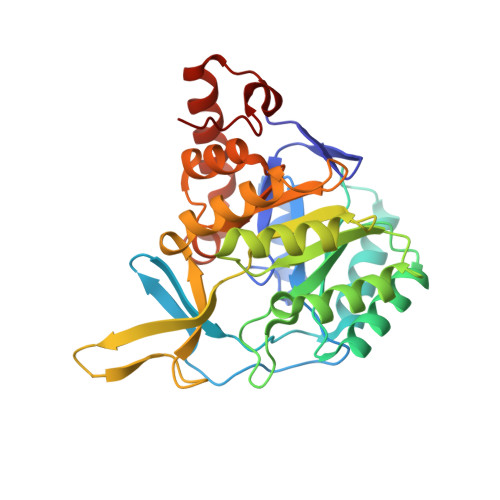
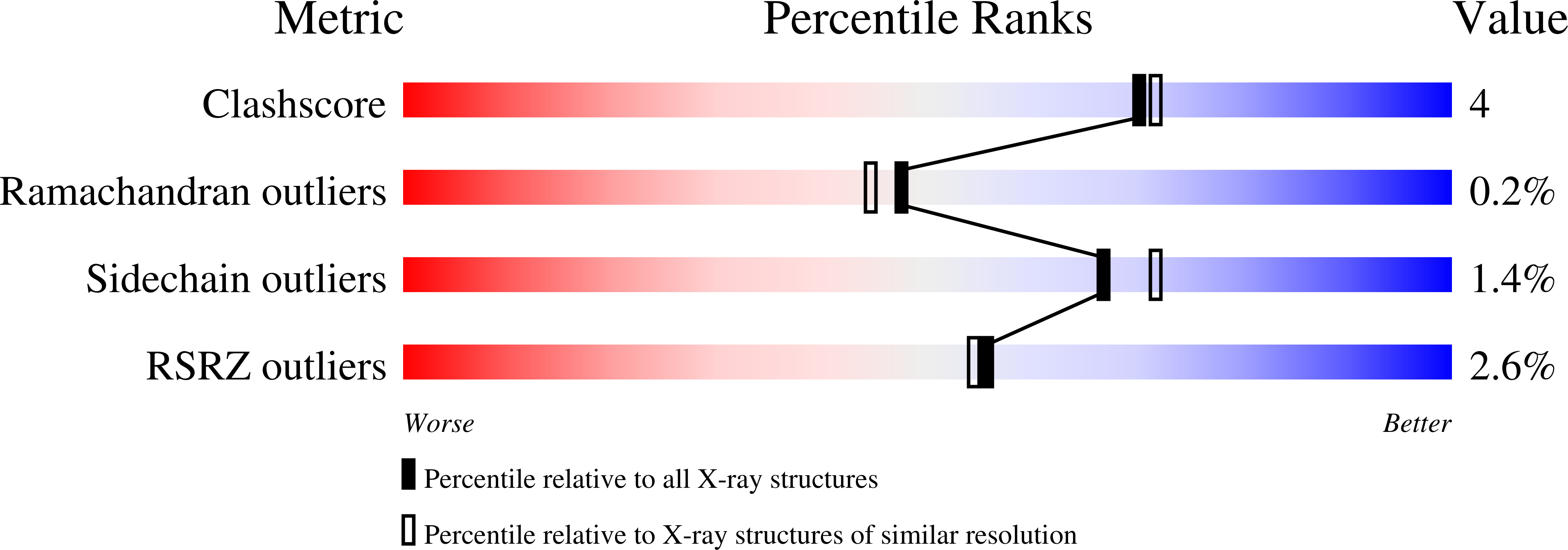The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis.
Rowland, P., Nielsen, F.S., Jensen, K.F., Larsen, S.(1997) Structure 5: 239-252
- PubMed: 9032071
- DOI: https://doi.org/10.1016/s0969-2126(97)00182-2
- Primary Citation of Related Structures:
1DOR - PubMed Abstract:
. Dihydroorotate dehydrogenase (DHOD) is a flavin mononucleotide containing enzyme, which catalyzes the oxidation of (S)-dihydroorotate to orotate, the fourth step in the de novo biosynthesis of pyrimidine nucleotides. Lactococcus lactis contains two genes encoding different functional DHODs whose sequences are only 30% identical. One of these enzymes, DHODA, is a highly efficient dimer, while the other, DHODB, shows optimal activity only in the presence of an iron-sulphur cluster containing protein with which it forms a complex tetramer. Sequence alignments have identified three different families among the DHODs: the two L. lactis enzymes belong to two of the families, whereas the enzyme from E. coli is a representative of the third. As no three-dimensional structures of DHODs are currently available, we set out to determine the crystal structure of DHODA from L. lactis. The differences between the two L. lactis enzymes make them particularly interesting for studying flavoprotein redox reactions and for identifying the differences between the enzyme families. . The crystal structure of DHODA has been determined to 2.0 resolution. The enzyme is a dimer of two crystallographically independent molecules related by a non-crystallographic twofold axis. The protein folds into and alpha/beta barrel with the flavin molecule sitting between the top of the barrel and a subdomain formed by several barrel inserts. Above the flavin isoalloxazine ring there is a small water filled cavity, completely buried beneath the protein surface and surrounded by many conserved residues. This cavity is proposed as the substrate-binding site. . The crystal structure has allowed the function of many of the conserved residues in DHODs to be identified: many of these are associated with binding the flavin group. Important differences were identified in some of the active-site residues which vary across the distinct DHOD families, implying significant mechanistic differences. The substrate cavity, although buried, is located beneath a highly conserved loop which is much less ordered than the rest of the protein and may be important in giving access to the cavity. The location of the conserved residues surrounding this cavity suggests the potential orientation of the substrate.
Organizational Affiliation:
Centre for Crystallographic Studies, Department of Chemistry University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen O, Denmark.