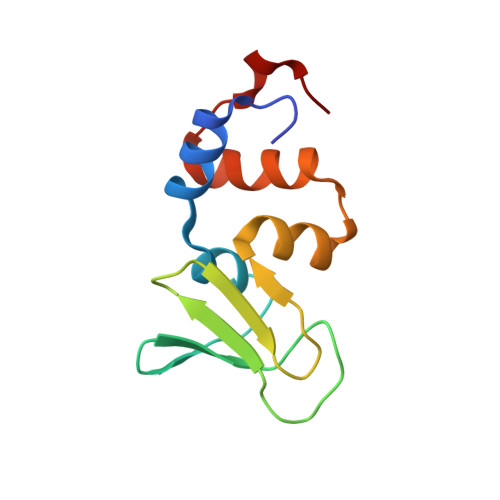
Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase.
Pan, H., Wigley, D.B.(2000) Structure 8: 231-239
- PubMed: 10745010
- DOI: https://doi.org/10.1016/s0969-2126(00)00101-5
- Primary Citation of Related Structures:
1D0Q - PubMed Abstract:
DNA primases catalyse the synthesis of the short RNA primers that are required for DNA replication by DNA polymerases. Primases comprise three functional domains: a zinc-binding domain that is responsible for template recognition, a polymerase domain, and a domain that interacts with the replicative helicase, DnaB. We present the crystal structure of the zinc-binding domain of DNA primase from Bacillus stearothermophilus, determined at 1.7 A resolution. This is the first high-resolution structural information about any DNA primase. A model is discussed for the interaction of this domain with the single-stranded DNA template. The structure of the DNA primase zinc-binding domain confirms that the protein belongs to the zinc ribbon subfamily. Structural comparison with other nucleic acid binding proteins suggests that the beta sheet of primase is likely to be the DNA-binding surface, with conserved residues on this surface being involved in the binding and recognition of DNA.
Organizational Affiliation:
Sir William Dunn School of Pathology, University of Oxford, Oxford, OX1 3RE, UK.