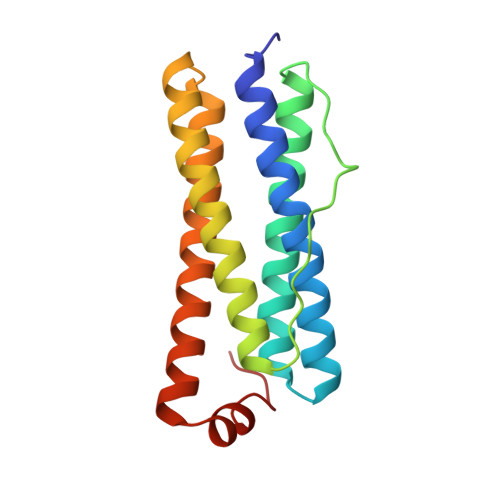
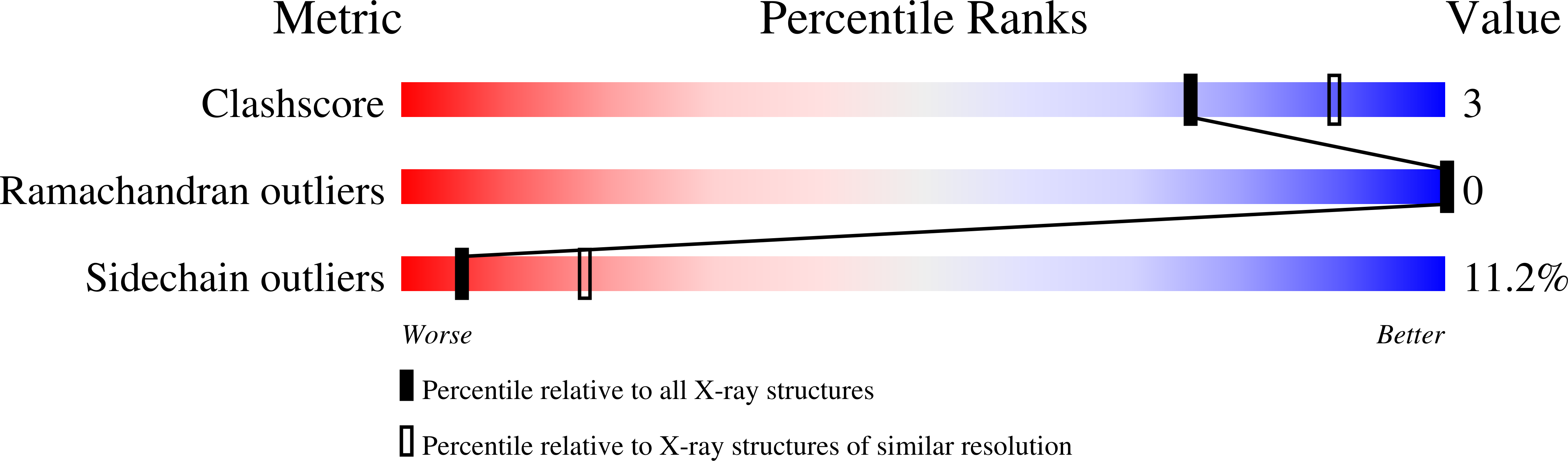Structure of a monoclinic crystal from of cyctochrome b1 (Bacterioferritin) from E. coli.
Dautant, A., Meyer, J.B., Yariv, J., Precigoux, G., Sweet, R.M., Kalb, A.J., Frolow, F.(1998) Acta Crystallogr D Biol Crystallogr 54: 16-24
- PubMed: 9867433
- DOI: https://doi.org/10.1107/s0907444997006811
- Primary Citation of Related Structures:
1BFR - PubMed Abstract:
Crystals of E. coli cytochrome b1, alias bacterioferritin, were grown fr om a low ionic strength solution. The resulting monoclniic P21 structure was solved by molecular replacement and refined using noncrystallographi c symmetries applied to the fundamental unit, consisting of two protein subunits and a single haem. From the Patterson self-rotation results it was shown that the asymmetric unit of the monoclinic crystal consists of 12 such dimers and corresponds to a complete, nearly spherical, molecule of bacterioferritin (M4 = 450 kDa) of 432 point-group symmetry. It is thus the most symmetrical cytochrome. As previously determined for the tetragonal form, the haem is located in a special position on a local twofold axis of the dimer. A bimetal centre is also observed within the four-helix bundle of each monomer; a metal-binding site is located on the fourfold axis.
Organizational Affiliation:
Unité Biophysique Structurale, ERS133 CNRS, Université de Bordeaux 1, France.