NMR Structure of the Natural Killer Cell Receptor 2B4: Implications for Ligand Recognition
Ames, J.B., Vyas, V., Lusin, J.D., Mariuzza, R.(2005) Biochemistry 44: 6416-6423
- PubMed: 15850375
- DOI: https://doi.org/10.1021/bi050139s
- Primary Citation of Related Structures:
1Z2K - PubMed Abstract:
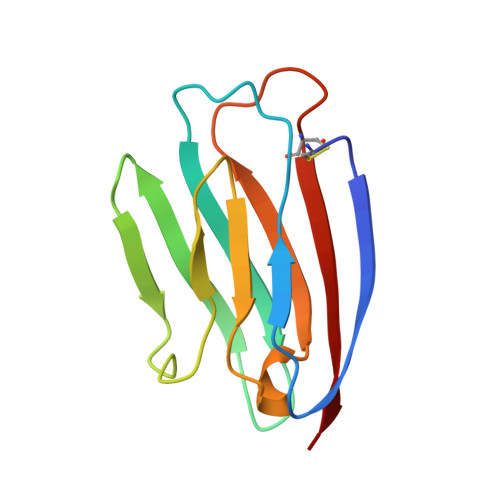
2B4, a transmembrane receptor expressed primarily on natural killer (NK) cells and on a subset of CD8(+) T cells, plays an important role in activating NK-mediated cytotoxicity through its interaction with CD48 on target cells. We report here the atomic-resolution structure of the ligand-binding (D1) domain of 2B4 in solution determined by nuclear magnetic resonance (NMR) spectroscopy. The overall main chain structure resembles an immunoglobulin variable (V) domain fold, very similar to that seen previously for domain 1 of CD2 and CD4. The structure contains nine beta-strands assembled into two beta-sheets conventionally labeled DEB and AGFCC'C' '. The six-stranded sheet (AGFCC'C' ') contains structural features that may have implications for ligand recognition and receptor function. A noncanonical disulfide bridge between Cys2 and Cys99 stabilizes a long and parallel beta-structure between strand A (residues 3-12) and strand G (residues 100-108). A beta-bulge at residues Glu45 and Ile46 places a bend in the middle of strand C' that orients two conserved and adjacent hydrophobic residues (Ile46 and Leu47) inside the beta-sandwich as seen in other V domains. Finally, the FG-loop (implicated in ligand recognition in the CD2-CD58 complex) is dynamically disordered in 2B4 in the absence of a ligand. We propose that ligand binding to 2B4 might stabilize the structure of the FG-loop in the ligand complex.
Organizational Affiliation:
Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, Rockville, Maryland 20850, USA. james@carb.nist.gov