A Third Specificity-Determining Site in Mu 2 Adaptin for Sequences Upstream of Yxx Phi Sorting Motifs
Owen, D.J., Setiadi, H., Evans, P.R., Mcever, R.P., Green, S.A.(2001) Traffic 2: 105
- PubMed: 11247301
- DOI: https://doi.org/10.1034/j.1600-0854.2001.020205.x
- Primary Citation of Related Structures:
1HES - PubMed Abstract:
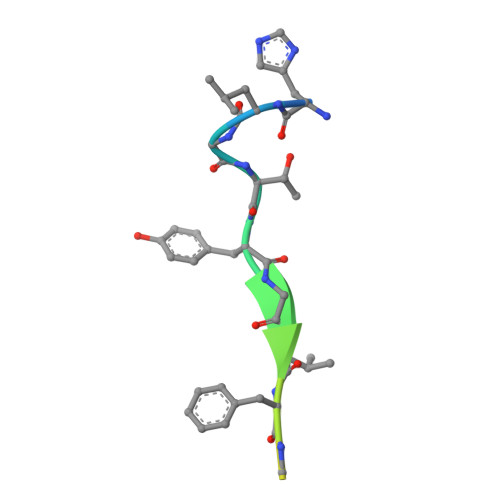
Internalization signals of the Yxx phi type (phi = bulky hydrophobic side chain) interact with the mu 2 chain of AP-2 adaptors. Internalization activity is intolerant of non-conservative substitution of either the tyrosine or the phi side chains, which bind to hydrophobic pockets in mu 2 adaptin in a conformation described as 'a two pinned plug into a socket'. P-selectin, a type I transmembrane protein, contains the Yxx phi-like sequence YGVF in its cytoplasmic domain, but substitution of either the tyrosine or phenylalanine with alanine in the full-length protein causes only small changes in the rate of endocytosis. It is shown here that the sequence YGVF contained within a peptide corresponding to the 17 COOH-terminal amino acids of P-selectin binds to mu 2 adaptin in the same fashion previously seen for other Yxx phi motifs. In addition, the P-selectin peptide binds to a third hydrophobic pocket in mu 2 adaptin through a leucine at position Y-3 in the peptide. This structure suggests that some sequences can function as a 'three pinned plug', in which internalization activity is not critically dependent on any one of the three interacting side chains.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.