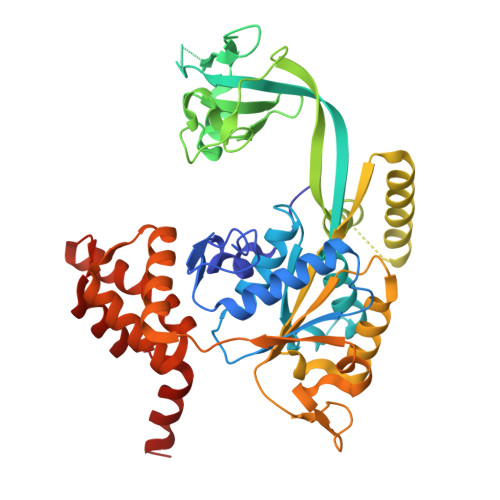
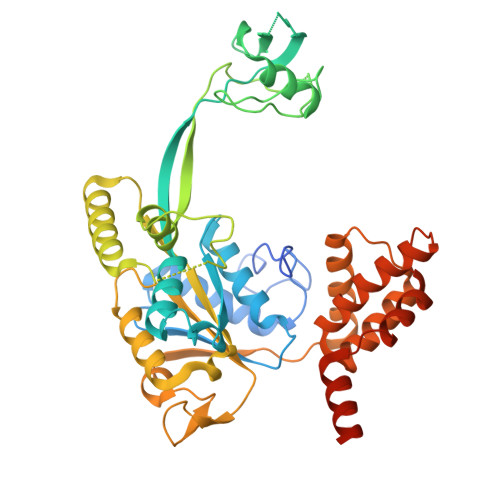
An integrative structural biology approach reveals the dynamic organization of the R2SP quaternary chaperone complex.
Santo, P.E., Chagot, M.E., Gizardin-Fredon, H., Ley, M., Chenuel, T., Desligniere, E., Plassart, L., Paiva, A.C.F., Sousa, P.M.F., Bertrand, E., Charpentier, B., Verheggen, C., Quinternet, M., Meyer, P., Bandeiras, T.M., Cianferani, S., Plisson-Chastang, C., Manival, X.(2026) Nat Commun
- PubMed: 41667496
- DOI: https://doi.org/10.1038/s41467-026-69157-x
- Primary Citation of Related Structures:
9HB4, 9HPO - PubMed Abstract:
R2SP belongs to the R2TP-like quaternary chaperone family and consists of RUVBL1/RUVBL2 AAA+ ATPases, which powers the machinery, and SPAG1 and PIH1D2 adapter proteins that engage specific clients to promote their quaternary assembly. However, little is known about the structure of R2SP and the precise mode of action of these R2TP-like complexes. Here, we combined biochemical (ATPase and fluorescence polarization assays) and structural approaches (NMR, structural mass spectrometry, cryo-EM) to investigate the 3D organization of the R2SP complex, its mode of assembly and ATPase activity. Together with our binding, mutational and kinetic studies, these results led us to propose a model in which SPAG1 and PIH1D2 bind and cooperatively engage RUVBL1/RUVBL2 to produce R2SP. This reveals a 3D structure close to the canonical R2TP complex but also highlights differences in RUVBL1/RUVBL2 ATPase activity, as well as the cooperative binding of SPAG1 and PIH1D2 to this catalytic core that may explain functional difference between the two systems.
- Centre de Biologie Intégrative (CBI), University of Toulouse CNRS, Equipe labellisée Ligue Nationale Contre le Cancer, Toulouse, France.
Organizational Affiliation: