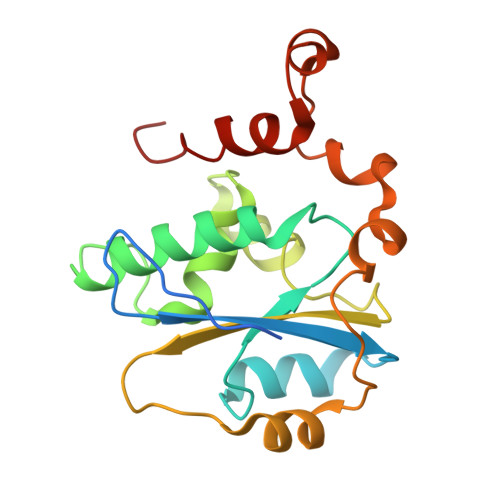
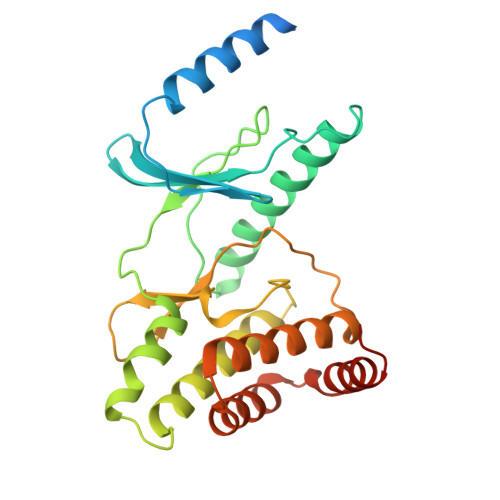
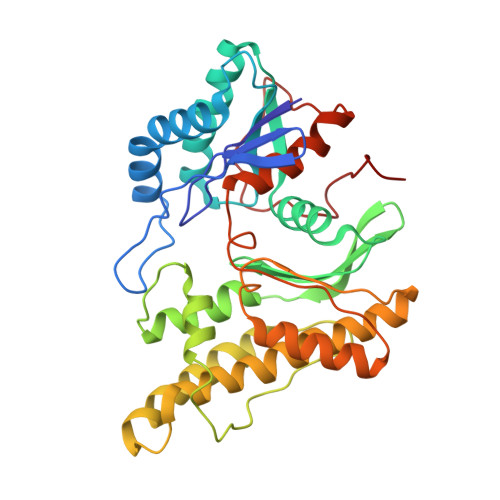
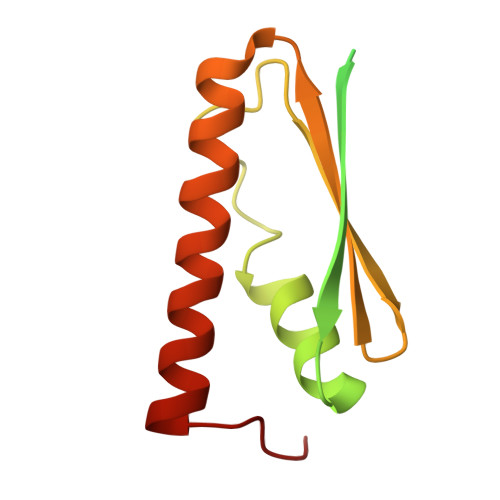
Cryo-EM structure of the human KEOPS complex
Cirio, C., Auxilien, S., Liger, D., Fernandes, C.A.H., Dammak, R., Venien-Bryan, C., Mollet, G., Zelie, E., Malhao, M., Arteni, A.A., Touboul, D., Antignac, C., Missoury, S., Van Tilbeurgh, H., Collinet, B.To be published.