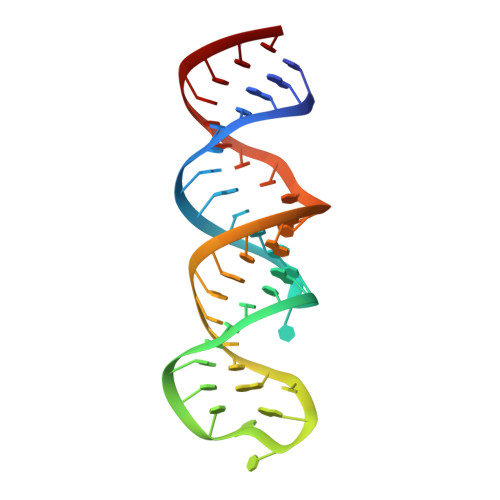
High-resolution structure of stem-loop 4 from the 5'-UTR of SARS-CoV-2 solved by solution state NMR.
Vogele, J., Hymon, D., Martins, J., Ferner, J., Jonker, H.R.A., Hargrove, A.E., Weigand, J.E., Wacker, A., Schwalbe, H., Wohnert, J., Duchardt-Ferner, E.(2023) Nucleic Acids Res 51: 11318-11331
- PubMed: 37791874
- DOI: https://doi.org/10.1093/nar/gkad762
- Primary Citation of Related Structures:
8CQ1 - PubMed Abstract:
We present the high-resolution structure of stem-loop 4 of the 5'-untranslated region (5_SL4) of the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) genome solved by solution state nuclear magnetic resonance spectroscopy. 5_SL4 adopts an extended rod-like structure with a single flexible looped-out nucleotide and two mixed tandem mismatches, each composed of a G•U wobble base pair and a pyrimidine•pyrimidine mismatch, which are incorporated into the stem-loop structure. Both the tandem mismatches and the looped-out residue destabilize the stem-loop structure locally. Their distribution along the 5_SL4 stem-loop suggests a role of these non-canonical elements in retaining functionally important structural plasticity in particular with regard to the accessibility of the start codon of an upstream open reading frame located in the RNA's apical loop. The apical loop-although mostly flexible-harbors residual structural features suggesting an additional role in molecular recognition processes. 5_SL4 is highly conserved among the different variants of SARS-CoV-2 and can be targeted by small molecule ligands, which it binds with intermediate affinity in the vicinity of the non-canonical elements within the stem-loop structure.
Organizational Affiliation:
Institute for Molecular Biosciences, Goethe-University Frankfurt, Max-von-Laue-Strasse 9, 60438 Frankfurt/M., Germany.