Three enigmatic BioH isoenzymes are programmed in the early stage of mycobacterial biotin synthesis, an attractive anti-TB drug target.
Xu, Y., Yang, J., Li, W., Song, S., Shi, Y., Wu, L., Sun, J., Hou, M., Wang, J., Jia, X., Zhang, H., Huang, M., Lu, T., Gan, J., Feng, Y.(2022) PLoS Pathog 18: e1010615-e1010615
- PubMed: 35816546
- DOI: https://doi.org/10.1371/journal.ppat.1010615
- Primary Citation of Related Structures:
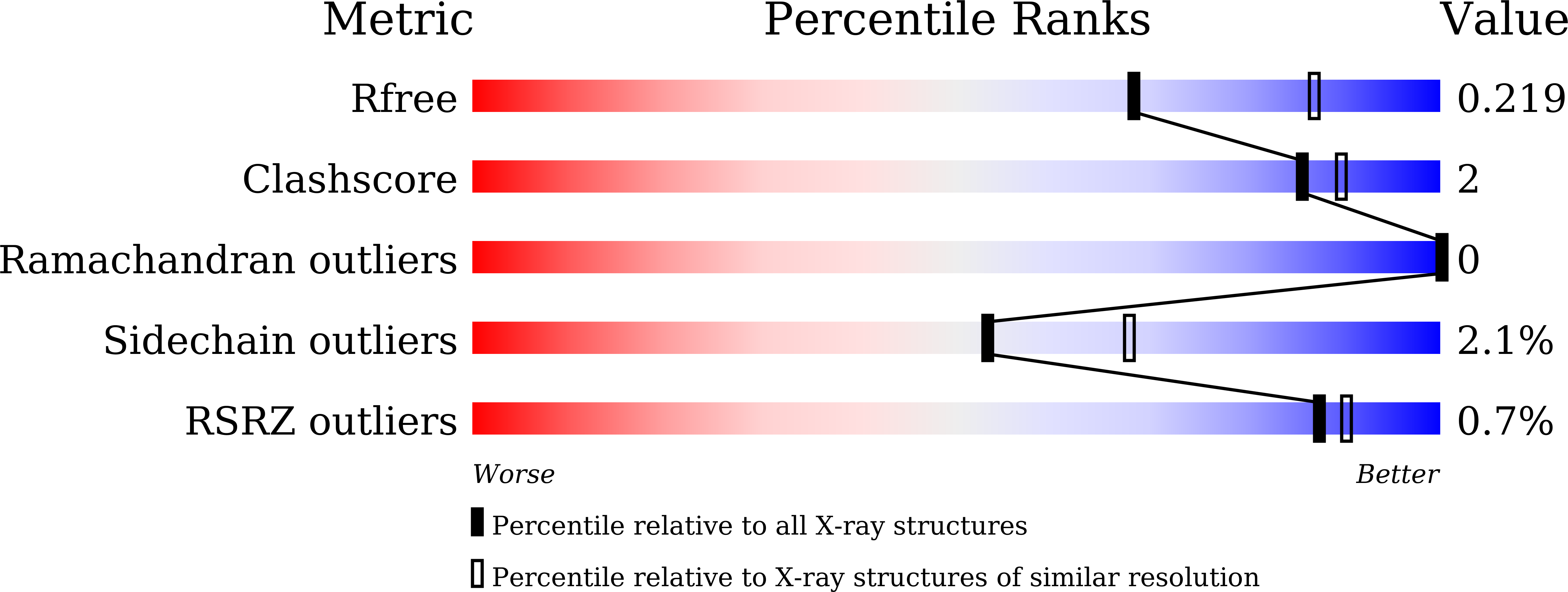
7WWF - PubMed Abstract:
Tuberculosis (TB) is one of the leading infectious diseases of global concern, and one quarter of the world's population are TB carriers. Biotin metabolism appears to be an attractive anti-TB drug target. However, the first-stage of mycobacterial biotin synthesis is fragmentarily understood. Here we report that three evolutionarily-distinct BioH isoenzymes (BioH1 to BioH3) are programmed in biotin synthesis of Mycobacterium smegmatis. Expression of an individual bioH isoform is sufficient to allow the growth of an Escherichia coli ΔbioH mutant on the non-permissive condition lacking biotin. The enzymatic activity in vitro combined with biotin bioassay in vivo reveals that BioH2 and BioH3 are capable of removing methyl moiety from pimeloyl-ACP methyl ester to give pimeloyl-ACP, a cognate precursor for biotin synthesis. In particular, we determine the crystal structure of dimeric BioH3 at 2.27Å, featuring a unique lid domain. Apart from its catalytic triad, we also dissect the substrate recognition of BioH3 by pimeloyl-ACP methyl ester. The removal of triple bioH isoforms (ΔbioH1/2/3) renders M. smegmatis biotin auxotrophic. Along with the newly-identified Tam/BioC, the discovery of three unusual BioH isoforms defines an atypical 'BioC-BioH(3)' paradigm for the first-stage of mycobacterial biotin synthesis. This study solves a long-standing puzzle in mycobacterial nutritional immunity, providing an alternative anti-TB drug target.
Organizational Affiliation:
Departments of Microbiology, and General Intensive Care Unit of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, The People's Republic of China.