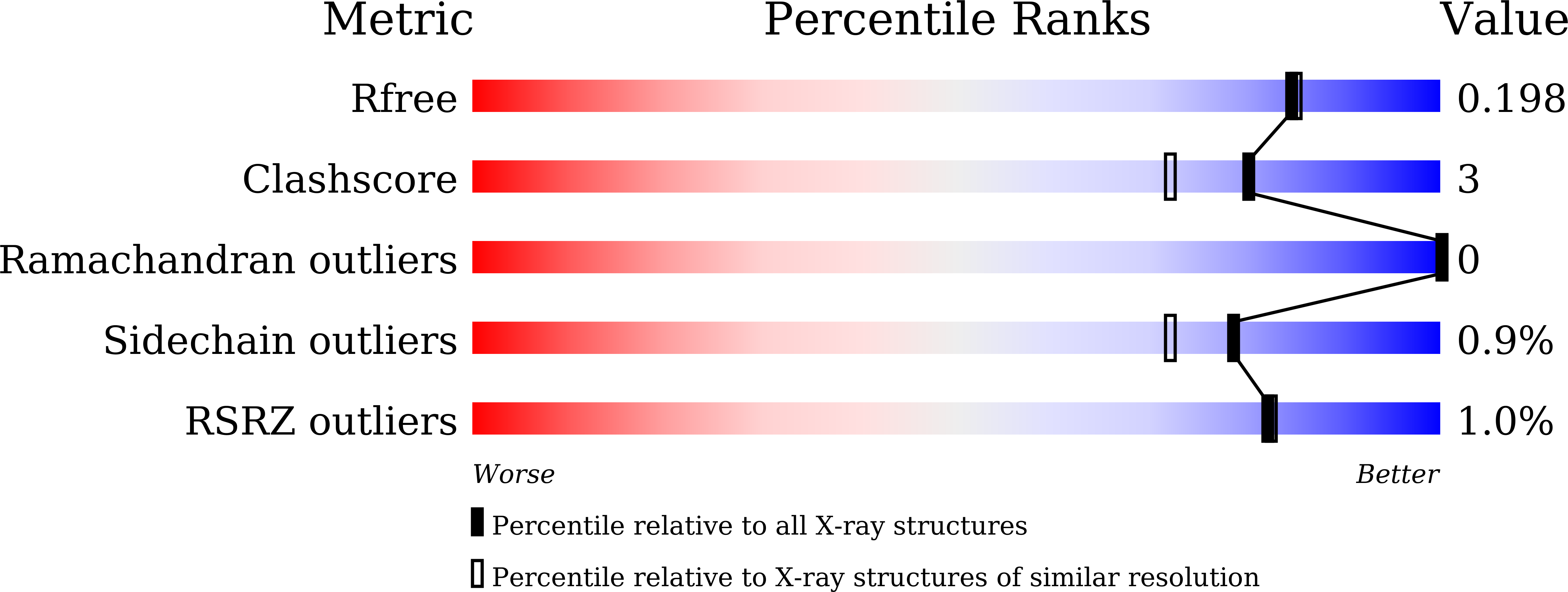
Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Zeng, W., Li, X., Yang, Y., Min, J., Huang, J.-W., Liu, W., Niu, D., Yang, X., Han, X., Zhang, L., Dai, L., Chen, C.-C., Guo, R.-T.(2022) ACS Catal 12: 3033-3040