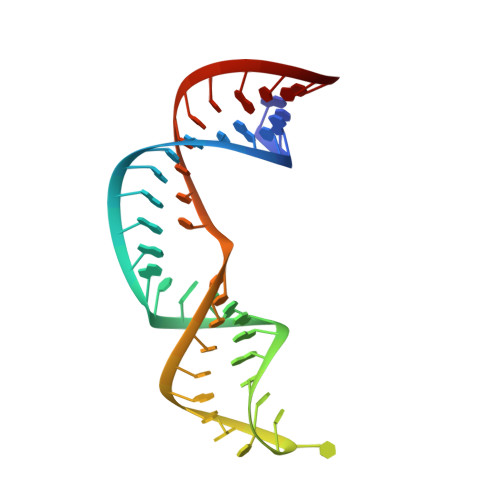
Solution Structure of NPSL2, A Regulatory Element in the oncomiR-1 RNA.
Liu, Y., Munsayac, A., Hall, I., Keane, S.C.(2022) J Mol Biol 434: 167688-167688
- PubMed: 35717998
- DOI: https://doi.org/10.1016/j.jmb.2022.167688
- Primary Citation of Related Structures:
7UGA - PubMed Abstract:
The miR-17 ∼ 92a polycistron, also known as oncomiR-1, is commonly overexpressed in multiple cancers and has several oncogenic properties. OncomiR-1 encodes six constituent microRNAs (miRs), each enzymatically processed with different efficiencies. However, the structural mechanism that regulates this differential processing remains unclear. Chemical probing of oncomiR-1 revealed that the Drosha cleavage sites of pri-miR-92a are sequestered in a four-way junction. NPSL2, an independent stem loop element, is positioned just upstream of pri-miR-92a and sequesters a crucial part of the sequence that constitutes the basal helix of pri-miR-92a. Disruption of the NPSL2 hairpin structure could promote the formation of a pri-miR-92a structure that is primed for processing by Drosha. Thus, NPSL2 is predicted to function as a structural switch, regulating pri-miR-92a processing. Here, we determined the solution structure of NPSL2 using solution NMR spectroscopy. This is the first high-resolution structure of an oncomiR-1 element. NPSL2 adopts a hairpin structure with a large, but highly structured, apical and internal loops. The 10-bp apical loop contains a pH-sensitive A + ·C mismatch. Additionally, several adenosines within the apical and internal loops have elevated pK a values. The protonation of these adenosines can stabilize the NPSL2 structure through electrostatic interactions. Our study provides fundamental insights into the secondary and tertiary structure of an important RNA hairpin proposed to regulate miR biogenesis.
Organizational Affiliation:
Biophysics Program, University of Michigan, 930 N. University Avenue, Ann Arbor, MI 48109, USA. Electronic address: https://twitter.com/YapingLiu5.