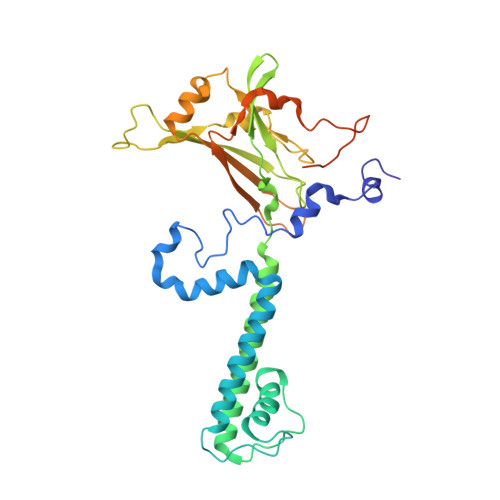
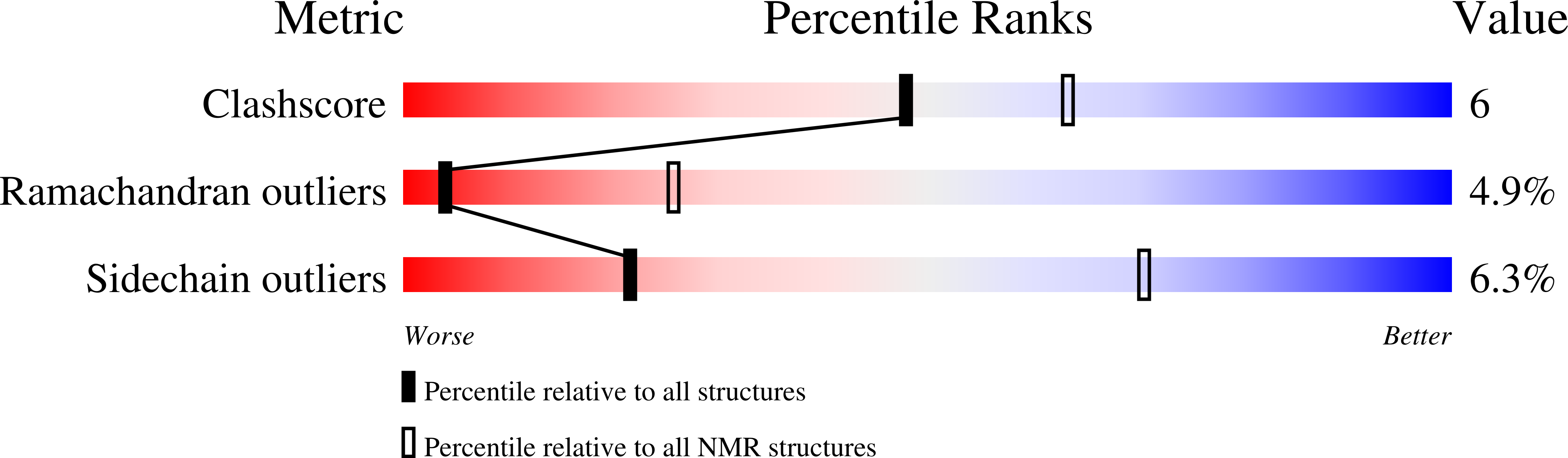Water Accessibility Refinement of the Extended Structure of KirBac1.1 in the Closed State.
Amani, R., Schwieters, C.D., Borcik, C.G., Eason, I.R., Han, R., Harding, B.D., Wylie, B.J.(2021) Front Mol Biosci 8: 772855-772855
- PubMed: 34917650
- DOI: https://doi.org/10.3389/fmolb.2021.772855
- Primary Citation of Related Structures:
7SWJ - PubMed Abstract:
NMR structures of membrane proteins are often hampered by poor chemical shift dispersion and internal dynamics which limit resolved distance restraints. However, the ordering and topology of these systems can be defined with site-specific water or lipid proximity. Membrane protein water accessibility surface area is often investigated as a topological function via solid-state NMR. Here we leverage water-edited solid-state NMR measurements in simulated annealing calculations to refine a membrane protein structure. This is demonstrated on the inward rectifier K + channel KirBac1.1 found in Burkholderia pseudomallei . KirBac1.1 is homologous to human Kir channels, sharing a nearly identical fold. Like many existing Kir channel crystal structures, the 1p7b crystal structure is incomplete, missing 85 out of 333 residues, including the N-terminus and C-terminus. We measure solid-state NMR water proximity information and use this for refinement of KirBac1.1 using the Xplor-NIH structure determination program. Along with predicted dihedral angles and sparse intra- and inter-subunit distances, we refined the residues 1-300 to atomic resolution. All structural quality metrics indicate these restraints are a powerful way forward to solve high quality structures of membrane proteins using NMR.
Organizational Affiliation:
Texas Tech University, Department of Chemistry and Biochemistry, Lubbock, TX, United States.