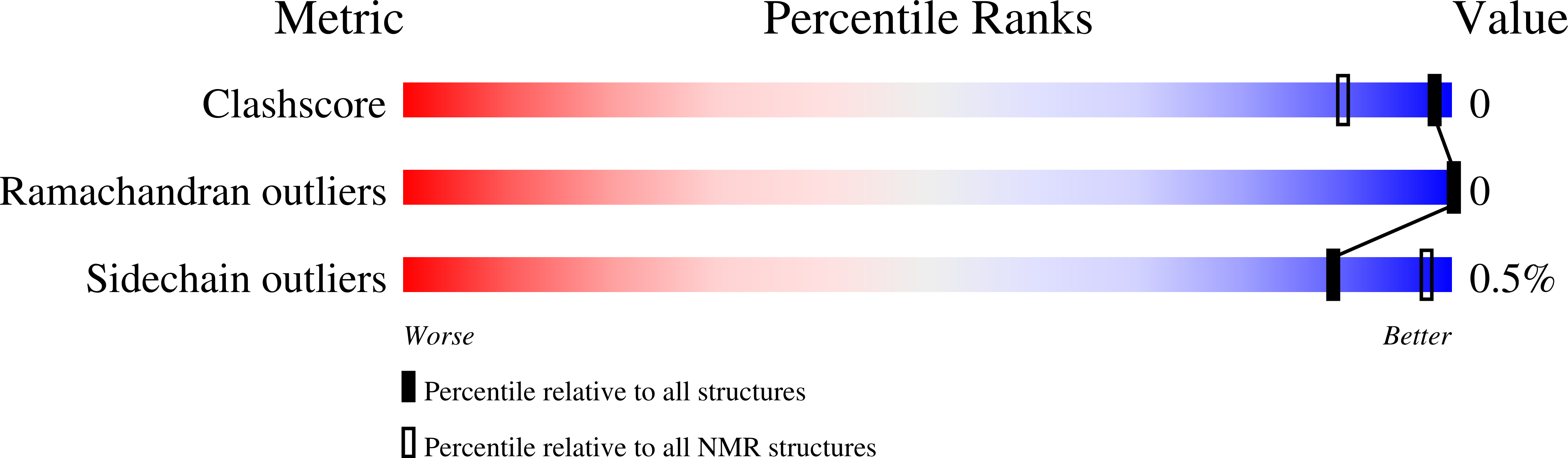Barrettides: A Peptide Family Specifically Produced by the Deep-Sea Sponge Geodia barretti .
Steffen, K., Laborde, Q., Gunasekera, S., Payne, C.D., Rosengren, K.J., Riesgo, A., Goransson, U., Cardenas, P.(2021) J Nat Prod 84: 3138-3146
- PubMed: 34874154
- DOI: https://doi.org/10.1021/acs.jnatprod.1c00938
- Primary Citation of Related Structures:
7SAG - PubMed Abstract:
Natural product discovery by isolation and structure elucidation is a laborious task often requiring ample quantities of biological starting material and frequently resulting in the rediscovery of previously known compounds. However, peptides are a compound class amenable to an alternative genomic, transcriptomic, and in silico discovery route by similarity searches of known peptide sequences against sequencing data. Based on the sequences of barrettides A and B, we identified five new barrettide sequences (barrettides C-G) predicted from the North Atlantic deep-sea demosponge Geodia barretti (Geodiidae). We synthesized, folded, and investigated one of the newly described barrettides, barrettide C (NVVPCFCVEDETSGAKTCIPDNCDASRGTNP, disulfide connectivity I-IV, II-III). Co-elution experiments of synthetic and sponge-derived barrettide C confirmed its native conformation. NMR spectroscopy and the anti-biofouling activity on larval settlement of the bay barnacle Amphibalanus improvisus (IC 50 0.64 μM) show that barrettide C is highly similar to barrettides A and B in both structure and function. Several lines of evidence suggest that barrettides are produced by the sponge itself and not one of its microbial symbionts.
Organizational Affiliation:
Pharmacognosy, Department of Pharmaceutical Biosciences, Biomedical Centre, Uppsala University, Husargatan 3, 751 23 Uppsala, Sweden.