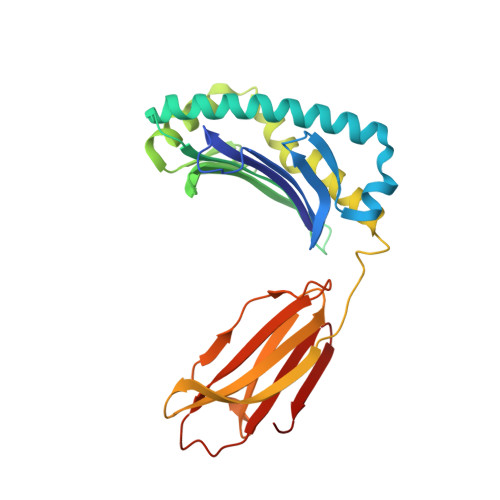
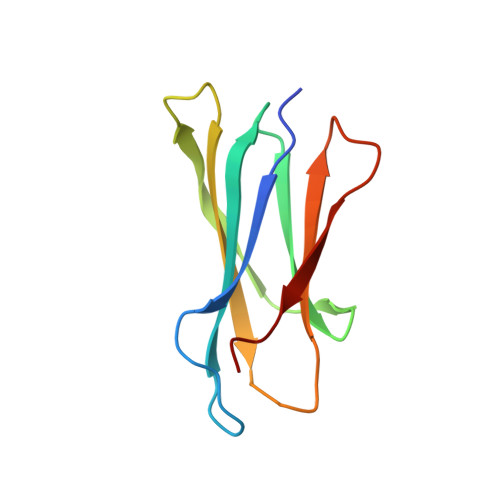
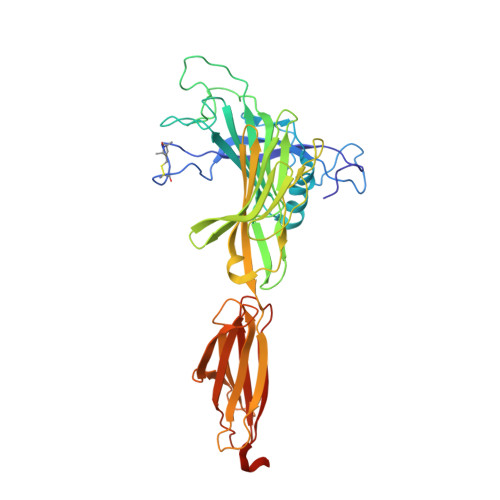
TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
McShan, A.C., Devlin, C.A., Papadaki, G.F., Sun, Y., Green, A.I., Morozov, G.I., Burslem, G.M., Procko, E., Sgourakis, N.G.(2022) Nat Chem Biol 18: 859-868
- PubMed: 35725941
- DOI: https://doi.org/10.1038/s41589-022-01049-9
- Primary Citation of Related Structures:
7RNO - PubMed Abstract:
Chaperones tapasin and transporter associated with antigen processing (TAP)-binding protein related (TAPBPR) associate with the major histocompatibility complex (MHC)-related protein 1 (MR1) to promote trafficking and cell surface expression. However, the binding mechanism and ligand dependency of MR1/chaperone interactions remain incompletely characterized. Here in vitro, biochemical and computational studies reveal that, unlike MHC-I, TAPBPR recognizes MR1 in a ligand-independent manner owing to the absence of major structural changes in the MR1 α 2-1 helix between empty and ligand-loaded molecules. Structural characterization using paramagnetic nuclear magnetic resonance experiments combined with restrained molecular dynamics simulations reveals that TAPBPR engages conserved surfaces on MR1 to induce similar adaptations to those seen in MHC-I/TAPBPR co-crystal structures. Finally, nuclear magnetic resonance relaxation dispersion experiments using 19 F-labeled diclofenac show that TAPBPR can affect the exchange kinetics of noncovalent metabolites with the MR1 groove, serving as a catalyst. Our results support a role of chaperones in stabilizing nascent MR1 molecules to enable loading of endogenous or exogenous cargo.
Organizational Affiliation:
Center for Computational and Genomic Medicine, Department of Pathology and Laboratory Medicine, The Children's Hospital of Philadelphia, Philadelphia, PA, USA.