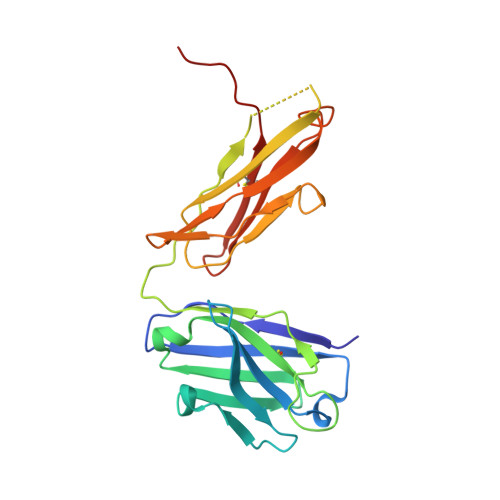
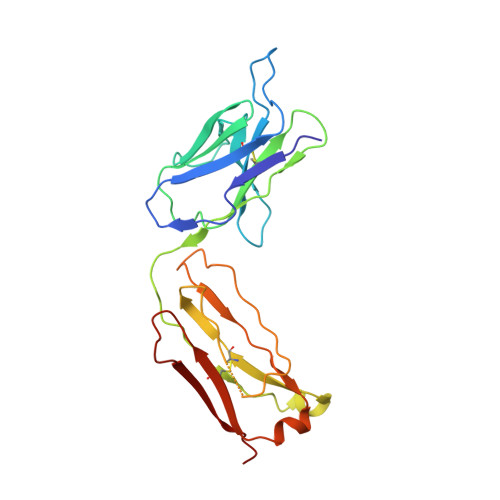
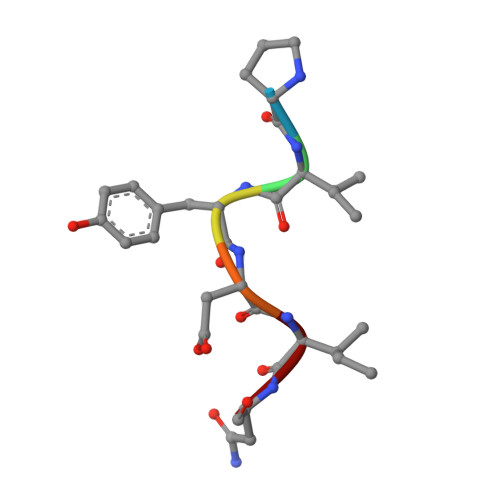
X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Govada, L., Saridakis, E., Kassen, S.C., Bin-Ramzi, A., Morgan, R.M., Chain, B., Helliwell, J.R., Chayen, N.E.(2021) IUCrJ 8: 678-683
- PubMed: 34258015
- DOI: https://doi.org/10.1107/S2052252521005340
- Primary Citation of Related Structures:
7NJZ, 7NW3 - PubMed Abstract:
C-C chemokine receptor 5 (CCR5) is a major co-receptor molecule used by HIV-1 to enter cells. This led to the hypothesis that stimulating an antibody response would block HIV with minimal toxicity. Here, X-ray crystallographic studies of the anti-CCR5 antibody RoAb13 together with two peptides were undertaken: one peptide is a 31-residue peptide containing the PIYDIN sequence and the other is the PIDYIN peptide alone, where PIYDIN is part of the N-terminal region of CCR5 previously shown to be important for HIV entry. In the presence of the longer peptide (the complete N-terminal domain), difference electron density was observed at a site within a hypervariable CDR3 binding region. In the presence of the shorter core peptide PIYDIN, difference electron density is again observed at this CDR3 site, confirming consistent binding for both peptides. This may be useful in the design of a new biomimetic to stimulate an antibody response to CCR5 in order to block HIV infection.
Organizational Affiliation:
Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, Faculty of Medicine, Sir Alexander Fleming Building, Imperial College London, London SW7 2AZ, United Kingdom.