A kink in DWORF helical structure controls the activation of the sarcoplasmic reticulum Ca 2+ -ATPase.
Reddy, U.V., Weber, D.K., Wang, S., Larsen, E.K., Gopinath, T., De Simone, A., Robia, S., Veglia, G.(2022) Structure 30: 360
- PubMed: 34875216
- DOI: https://doi.org/10.1016/j.str.2021.11.003
- Primary Citation of Related Structures:
7MPA - PubMed Abstract:
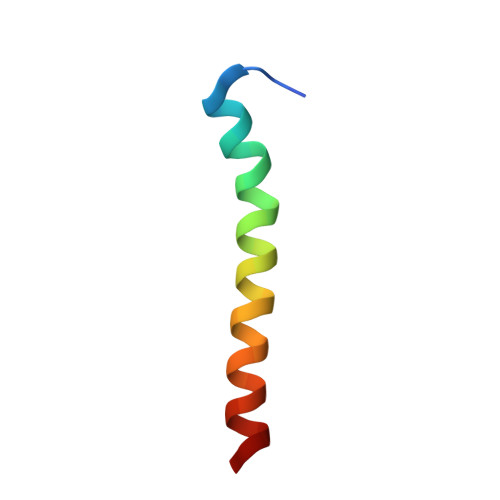
SERCA is a P-type ATPase embedded in the sarcoplasmic reticulum and plays a central role in muscle relaxation. SERCA's function is regulated by single-pass membrane proteins called regulins. Unlike other regulins, dwarf open reading frame (DWORF) expressed in cardiac muscle has a unique activating effect. Here, we determine the structure and topology of DWORF in lipid bilayers using a combination of oriented sample solid-state NMR spectroscopy and replica-averaged orientationally restrained molecular dynamics. We found that DWORF's structural topology consists of a dynamic N-terminal domain, an amphipathic juxtamembrane helix that crosses the lipid groups at an angle of 64°, and a transmembrane C-terminal helix with an angle of 32°. A kink induced by Pro15, unique to DWORF, separates the two helical domains. A single Pro15Ala mutant significantly decreases the kink and eliminates DWORF's activating effect on SERCA. Overall, our findings directly link DWORF's structural topology to its activating effect on SERCA.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, 6-155 Jackson Hall, Minneapolis, MN 55455, USA.