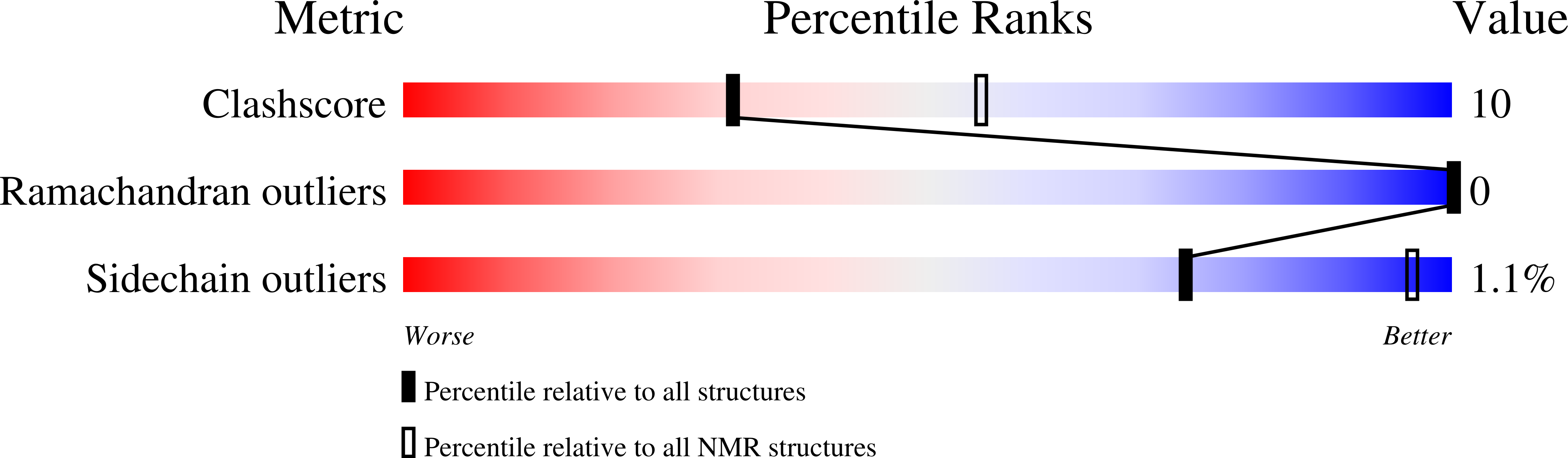Evolution of fold switching in a metamorphic protein.
Dishman, A.F., Tyler, R.C., Fox, J.C., Kleist, A.B., Prehoda, K.E., Babu, M.M., Peterson, F.C., Volkman, B.F.(2021) Science 371: 86-90
- PubMed: 33384377
- DOI: https://doi.org/10.1126/science.abd8700
- Primary Citation of Related Structures:
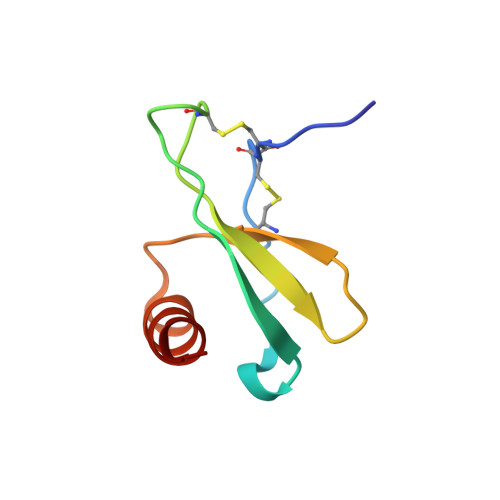
7JH1 - PubMed Abstract:
Metamorphic proteins switch between different folds, defying the protein folding paradigm. It is unclear how fold switching arises during evolution. With ancestral reconstruction and nuclear magnetic resonance, we studied the evolution of the metamorphic human protein XCL1, which has two distinct folds with different functions, making it an unusual member of the chemokine family, whose members generally adopt one conserved fold. XCL1 evolved from an ancestor with the chemokine fold. Evolution of a dimer interface, changes in structural constraints and molecular strain, and alteration of intramolecular protein contacts drove the evolution of metamorphosis. Then, XCL1 likely evolved to preferentially populate the noncanonical fold before reaching its modern-day near-equal population of folds. These discoveries illuminate how one sequence has evolved to encode multiple structures, revealing principles for protein design and engineering.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI, USA.