Microcrystal preparation for serial femtosecond X-ray crystallography of bacterial copper amine oxidase
Murakawa, T., Suzuki, M., Arima, T., Sugahara, M., Tanaka, T., Tanaka, R., Iwata, S., Nango, E., Tono, K., Hayashi, H., Fukui, K., Yano, T., Tanizawa, K., Okajima, T.(2021) Acta Crystallogr F Struct Biol Commun 77: 356-363
- PubMed: 34605440
- DOI: https://doi.org/10.1107/S2053230X21008967
- Primary Citation of Related Structures:
7F8K - PubMed Abstract:
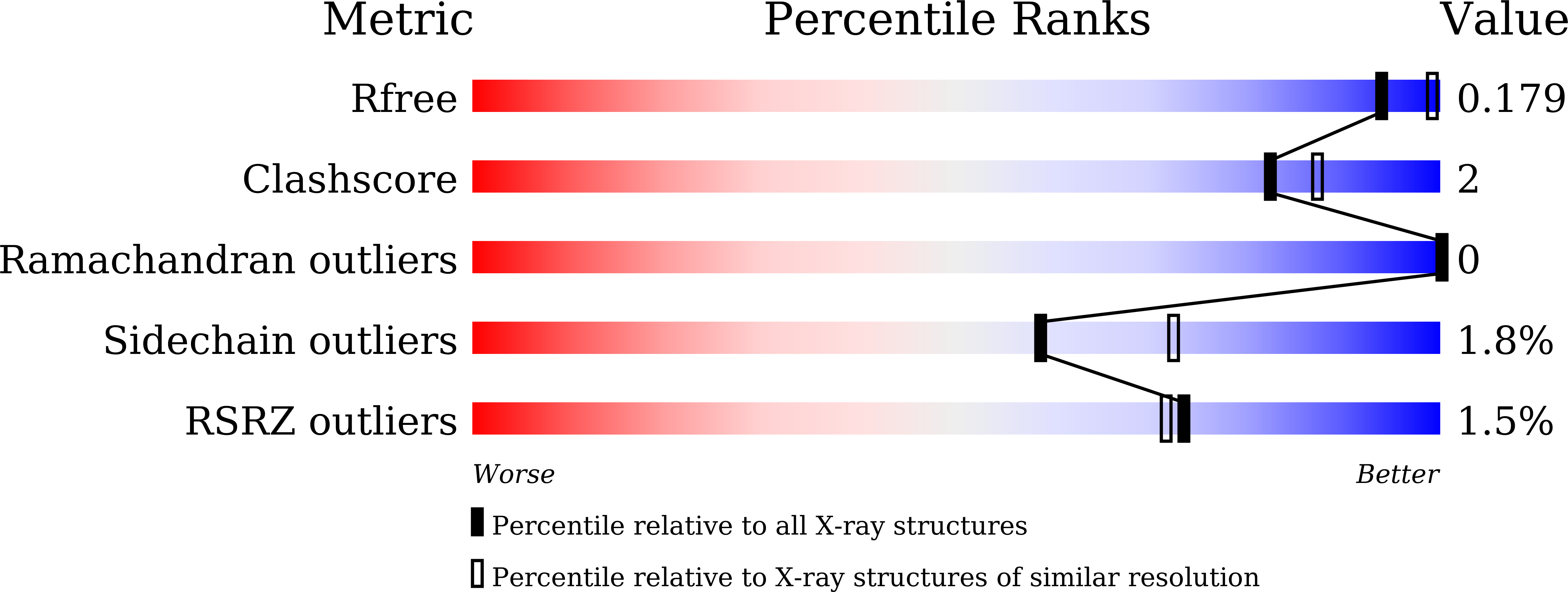
Recent advances in serial femtosecond X-ray crystallography (SFX) using X-ray free-electron lasers have paved the way for determining radiation-damage-free protein structures under nonfreezing conditions. However, the large-scale preparation of high-quality microcrystals of uniform size is a prerequisite for SFX, and this has been a barrier to its widespread application. Here, a convenient method for preparing high-quality microcrystals of a bacterial quinoprotein enzyme, copper amine oxidase from Arthrobacter globiformis, is reported. The method consists of the mechanical crushing of large crystals (5-15 mm 3 ), seeding the crushed crystals into the enzyme solution and standing for 1 h at an ambient temperature of ∼26°C, leading to the rapid formation of microcrystals with a uniform size of 3-5 µm. The microcrystals diffracted X-rays to a resolution beyond 2.0 Å in SFX measurements at the SPring-8 Angstrom Compact Free Electron Laser facility. The damage-free structure determined at 2.2 Å resolution was essentially identical to that determined previously by cryogenic crystallography using synchrotron X-ray radiation.
Organizational Affiliation:
Department of Biochemistry, Faculty of Medicine, Osaka Medical and Pharmaceutical University, 2-7 Daigakumachi, Takatsuki, Osaka 569-8686, Japan.