Positive Charge Introduction on the Surface of Thermostabilized PET Hydrolase Facilitates PET Binding and Degradation.
Nakamura, A.(2021) ACS Catal
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2021) ACS Catal
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LipIAF5-2 | 294 | uncultured bacterium | Mutation(s): 7 Gene Names: lipIAF5-2 | 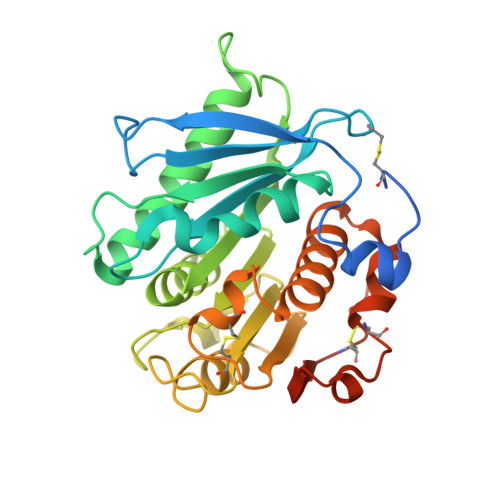 | |
UniProt | |||||
Find proteins for C3RYL0 (uncultured bacterium) Explore C3RYL0 Go to UniProtKB: C3RYL0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C3RYL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.24 | α = 90 |
| b = 86.72 | β = 90 |
| c = 46.21 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |