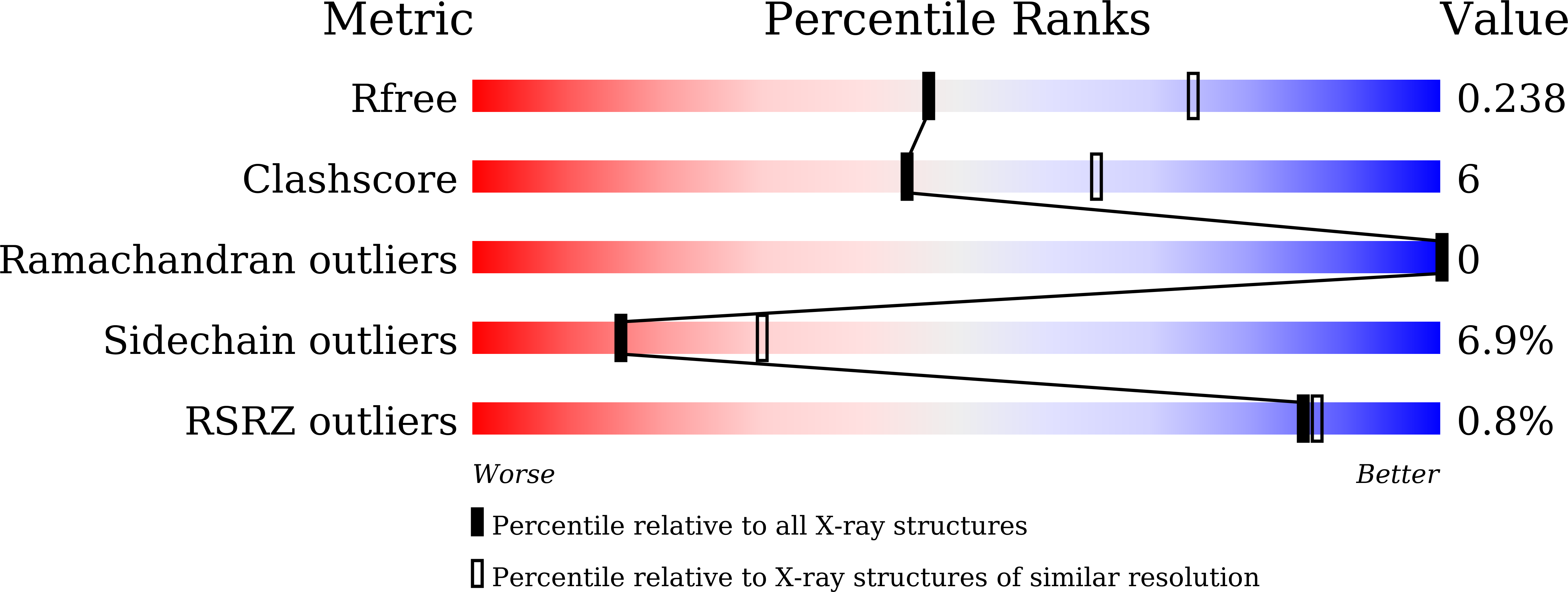
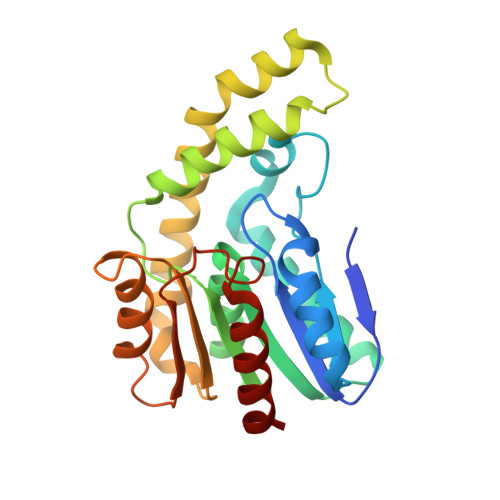
A reverse catalytic triad Asp containing loop shaping a wide substrate binding pocket of a feruloyl esterase from Lactobacillus plantarum.
Zhang, H., Wen, B., Liu, Y., Du, G., Wei, X., Imam, K.M.S.U., Zhou, H., Fan, S., Wang, F., Wang, Y., Xin, F.(2021) Int J Biol Macromol 184: 92-100
- PubMed: 34116094
- DOI: https://doi.org/10.1016/j.ijbiomac.2021.06.033
- Primary Citation of Related Structures:
7EBO - PubMed Abstract:
Feruloyl esterase is an indispensable biocatalyst in food processing, pesticide and pharmaceutical industries, catalyzing the cleavage of the ester bond cross-linked between the polysaccharide side chain of hemicellulose and ferulic acid in plant cell walls. LP_0796 from Lactobacillus plantarum was identified as a feruloyl esterase that may have potential applications in the food industry, but the lack of the substrate recognition and catalytic mechanisms limits its application. Here, LP_0796 showed the highest activity towards methyl caffeate at pH 6.6 and 40 °C. The crystal structure of LP_0796 was determined at 2.5 Å resolution and featured a catalytic triad Asp195-containing loop facing the opposite direction, thus forming a wider substrate binding pocket. Molecular docking simulation and site-directed mutagenesis studies further demonstrated that in addition to the catalytic triad (Ser94, Asp195, His225), Arg125 and Val128 played essential roles in the function of the active site. Our data also showed that Asp mutation of Ala23 and Ile198 increased the catalytic efficiency to 4- and 5-fold, respectively. Collectively, this work provided a better understanding of the substrate recognition and catalytic mechanisms of LP_0796 and may facilitate the future protein design of this important feruloyl esterase.
Organizational Affiliation:
Laboratory of Biomanufacturing and Food Engineering, Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, Beijing 100193, China.