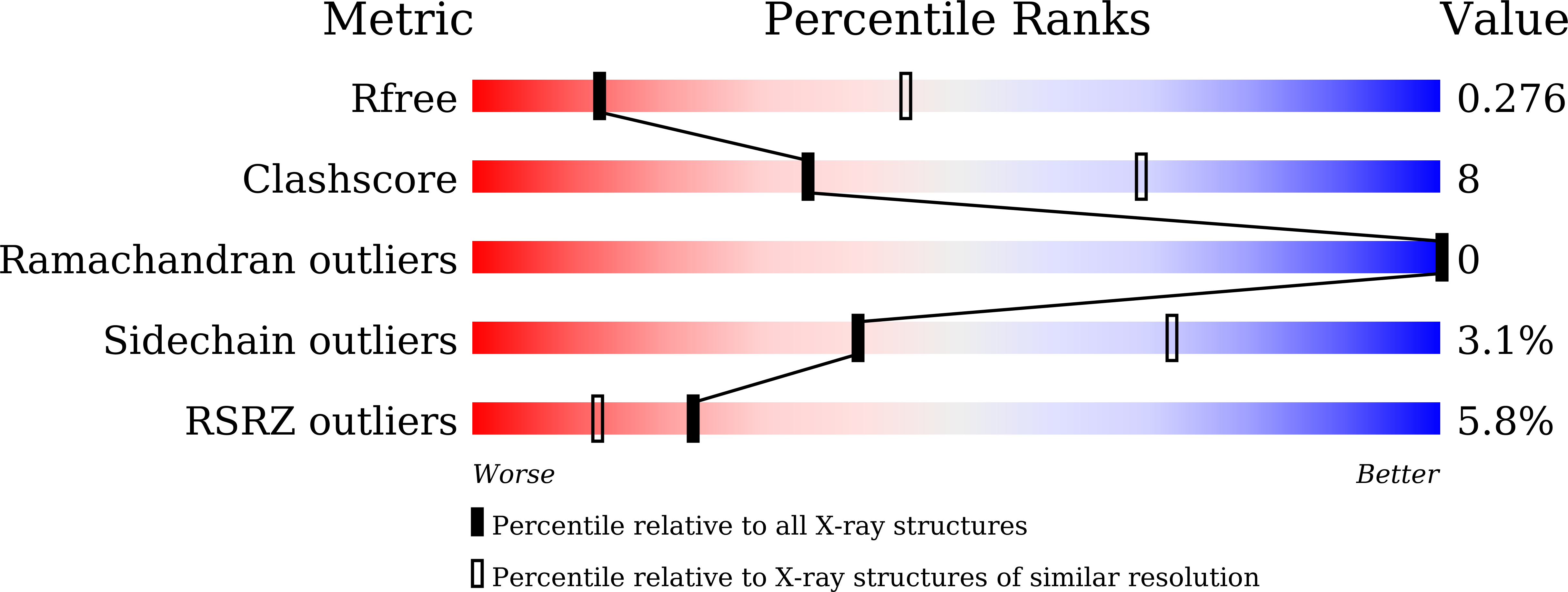
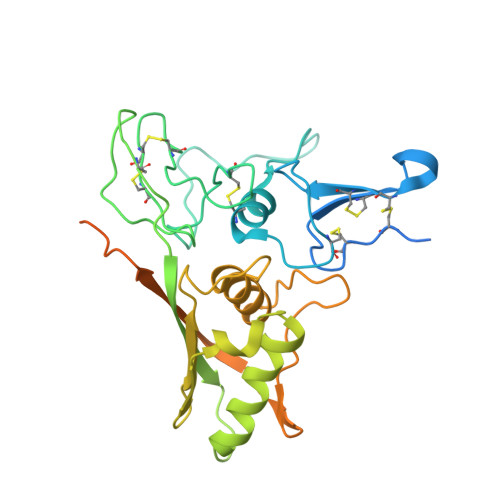
Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Sun, M., Zhang, H., Jiang, M., Chai, Y., Qi, J., Gao, G.F., Tan, S.(2021) iScience 24: 103190-103190
- PubMed: 34693228
- DOI: https://doi.org/10.1016/j.isci.2021.103190
- Primary Citation of Related Structures:
7E5M, 7E5N - PubMed Abstract:
Human trophoblast cell surface antigen 2 (TROP-2) is an important target of tumor therapy, and antibody-drug conjugates with sacituzumab targeting TROP-2 have been approved for the treatment of triple-negative breast cancer. Here, we report the crystal structures of TROP-2-ECD, which can be either cis- or trans- dimers depending on which distinct but overlapping interfaces is used to engage with monomers. The cis- or trans -tetrameric forms of TROP-2 can also be assembled with a non-overlapping interface with either cis - or trans- dimerization, suggesting that cis - and trans- dimers cluster on the cell surface. The binding site of sacituzumab on TROP-2 is mapped to be located on a stretched polypeptide in CPD (Q237-Q252), which is not involved in either cis- or trans- interactions. The present findings will improve understanding of the molecular assembly of TROP-2 on tumor cells and shed light on future design of biologics for tumor therapy.
Organizational Affiliation:
Research Network of Immunity and Health (RNIH), Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, China.