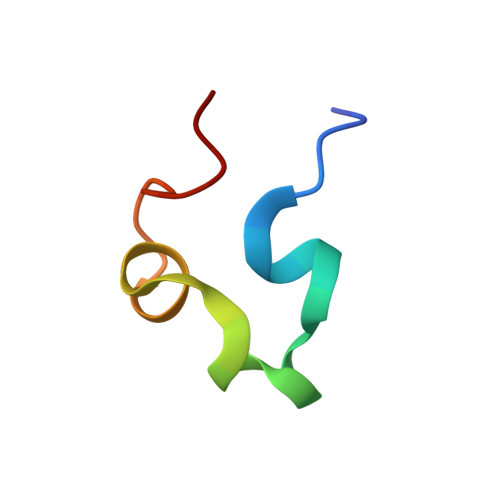
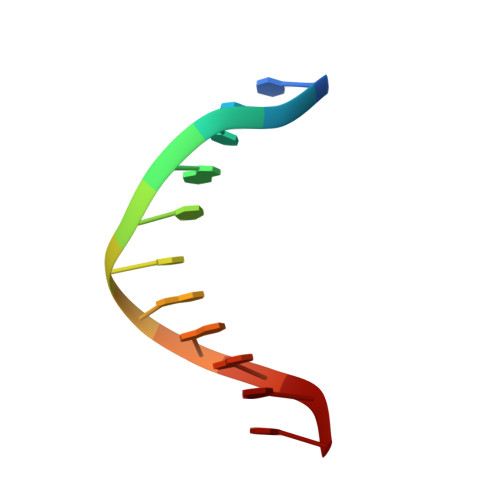
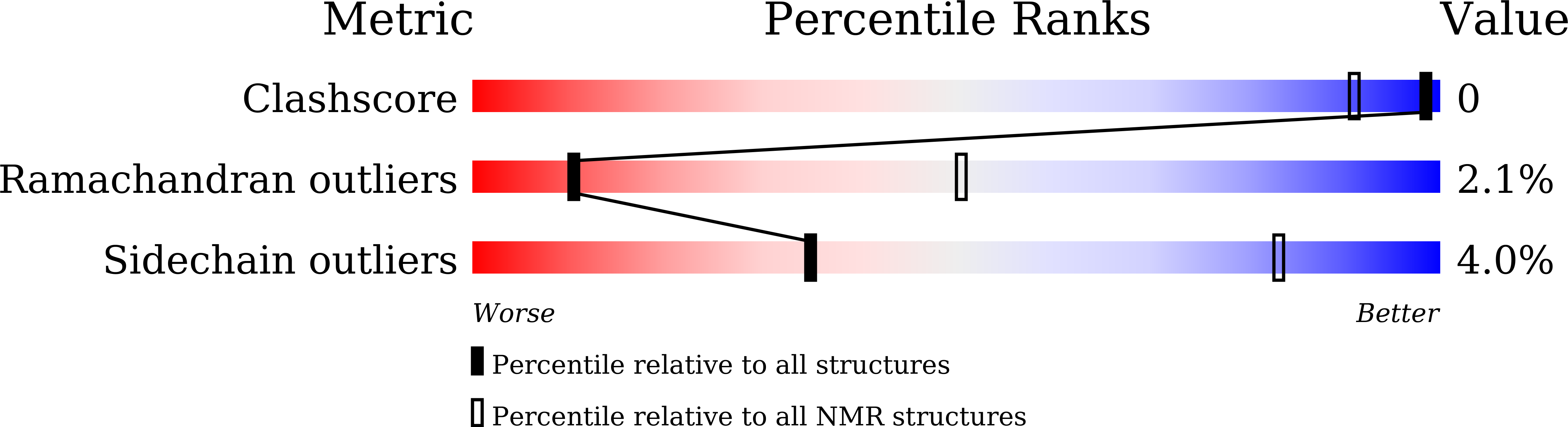Selective Nonmethylated CpG DNA Recognition Mechanism of Cysteine Clamp Domains.
Duan, B., Fu, D., Zhang, C., Ding, P., Dong, X., Xia, B.(2021) J Am Chem Soc 143: 7688-7697
- PubMed: 33983734
- DOI: https://doi.org/10.1021/jacs.1c00599
- Primary Citation of Related Structures:
7DTA - PubMed Abstract:
Methylation of DNA at CpG sites is a major mark for epigenetic regulation, but how transcription factors are influenced by CpG methylation is not well understood. Here, we report the molecular mechanisms of how the TCF (T-cell factor) and GEF (glucose transporter 4 enhancer factor) families of proteins selectively target unmethylated DNA sequences with a C-clamp type zinc finger domain. The structure of the C-clamp domain from human GEF family protein HDBP1 (C-clamp HDBP1 ) in complex with DNA was determined using NMR spectroscopy, which adopts a unique zinc finger fold and selectively binds RCCGG (R = A/G) DNA sequences with an "Arg···Trp-Lys-Lys" DNA recognition motif inserted in the major groove. The CpG base pairs are central to the binding due to multiple hydrogen bonds formed with the backbone carbonyl groups of Trp378 and Lys379, as well as the side chain ε-amino groups of Lys379 and Lys380 from C-clamp HDBP1 . Consequently, methylation of the CpG dinucleotide almost abolishes the binding. Homology modeling reveals that the C-clamp domain from human TCF1E (C-clamp TCF1E ) binds DNA through essentially the same mechanism, with a similar "Arg···Arg-Lys-Lys" DNA recognition motif. The substitution of tryptophan by arginine makes C-clamp HDBP1 prefer RCCGC DNA sequences. The two signature DNA recognition motifs are invariant in the GEF and TCF families of proteins, respectively, from fly to human. The recognition of the CpG dinucleotide through two consecutive backbone carbonyl groups is the same as that of the CXXC type unmethylated CpG DNA binding domains, suggesting a common mechanism shared by unmethylated CpG binding proteins.
Organizational Affiliation:
Beijing Nuclear Magnetic Resonance Center, College of Chemistry and Molecular Engineering, and School of Life Sciences, Peking University, Beijing 100871, China.