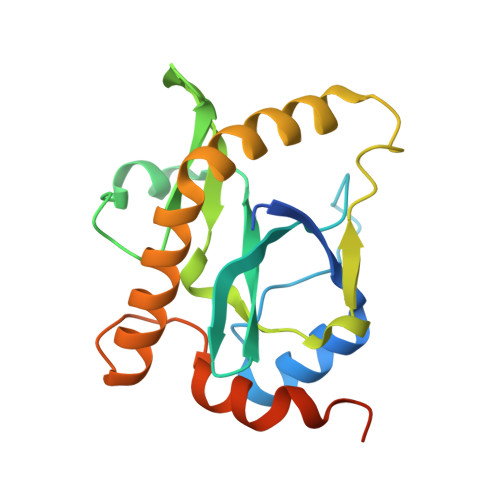
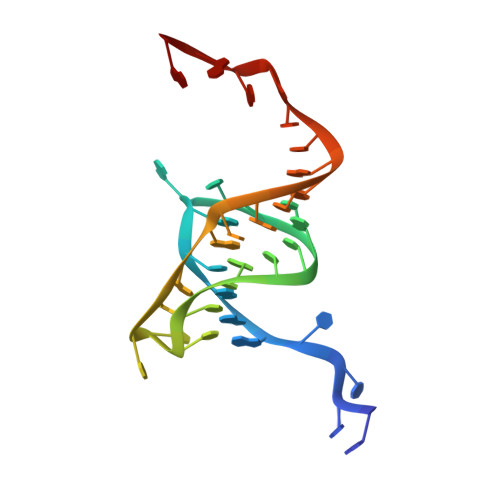
Identification, functional characterization, assembly and structure of ToxIN type III toxin-antitoxin complex from E. coli.
Manikandan, P., Sandhya, S., Nadig, K., Paul, S., Srinivasan, N., Rothweiler, U., Singh, M.(2022) Nucleic Acids Res 50: 1687-1700
- PubMed: 35018473
- DOI: https://doi.org/10.1093/nar/gkab1264
- Primary Citation of Related Structures:
7D8O - PubMed Abstract:
Toxin-antitoxin (TA) systems are proposed to play crucial roles in bacterial growth under stress conditions such as phage infection. The type III TA systems consist of a protein toxin whose activity is inhibited by a noncoding RNA antitoxin. The toxin is an endoribonuclease, while the antitoxin consists of multiple repeats of RNA. The toxin assembles with the individual antitoxin repeats into a cyclic complex in which the antitoxin forms a pseudoknot structure. While structure and functions of some type III TA systems are characterized, the complex assembly process is not well understood. Using bioinformatics analysis, we have identified type III TA systems belonging to the ToxIN family across different Escherichia coli strains and found them to be clustered into at least five distinct clusters. Furthermore, we report a 2.097 Å resolution crystal structure of the first E. coli ToxIN complex that revealed the overall assembly of the protein-RNA complex. Isothermal titration calorimetry experiments showed that toxin forms a high-affinity complex with antitoxin RNA resulting from two independent (5' and 3' sides of RNA) RNA binding sites on the protein. These results further our understanding of the assembly of type III TA complexes in bacteria.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bengaluru 560012, India.