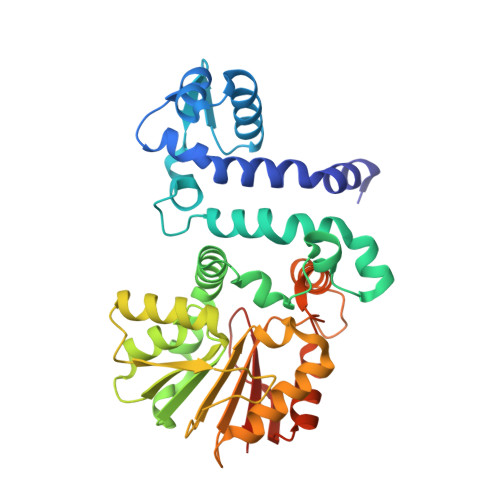
Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Qiu, S., Xu, D., Xu, M., Zhou, H., Sun, N., Zhang, L., Zhao, M., He, J., Ran, T., Sun, B., Wang, W.(2022) IUCrJ 9: 316-327