Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
Senga, A., Numoto, N., Yamashita, M., Iida, A., Ito, N., Kawai, F., Oda, M.(2021) J Biochem 169: 207-213
- PubMed: 32882044
- DOI: https://doi.org/10.1093/jb/mvaa102
- Primary Citation of Related Structures:
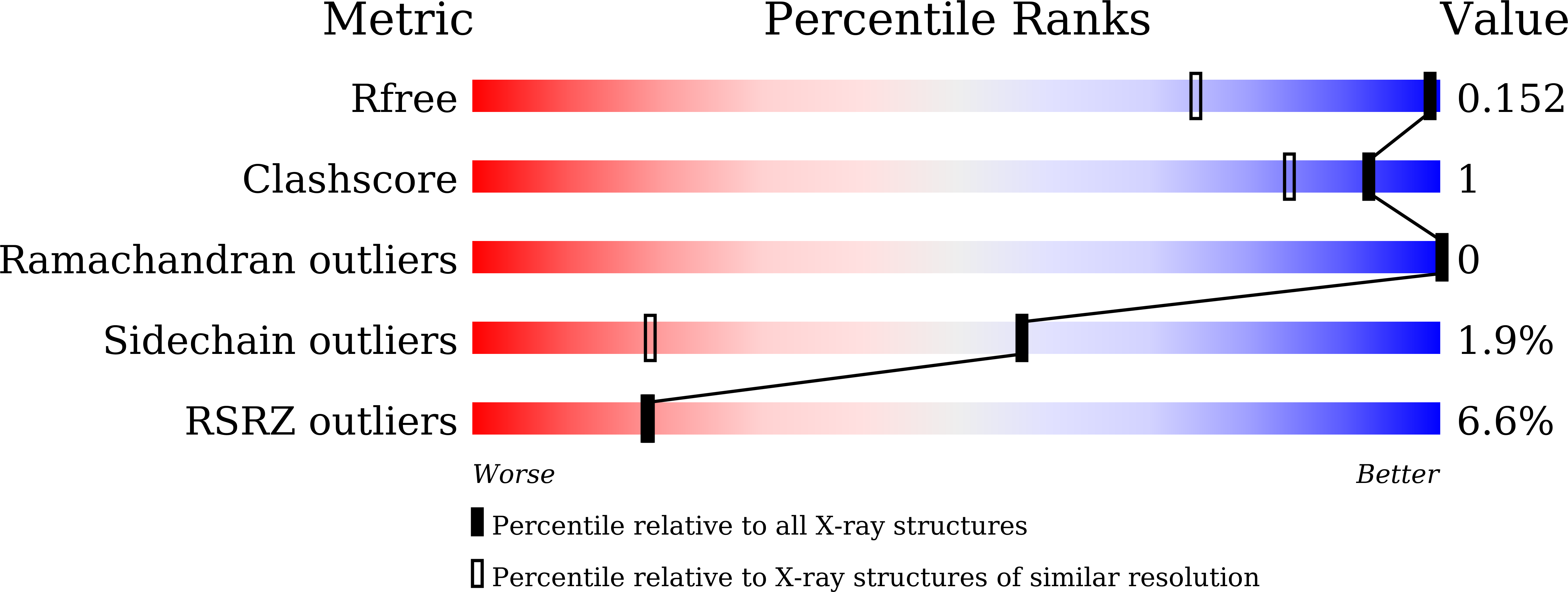
7CEF, 7CEH - PubMed Abstract:
An enzyme, Cut190, from a thermophilic isolate, Saccharomonospora viridis AHK190 could depolymerize polyethylene terephthalate (PET). The catalytic activity and stability of Cut190 and its S226P/R228S mutant, Cut190*, are regulated by Ca2+ binding. We previously determined the crystal structures of the inactive mutant of Cut190*, Cut190*S176A, in complex with metal ions, Ca2+ and Zn2+, and substrates, monoethyl succinate and monoethyl adipate. In this study, we determined the crystal structures of another mutant of Cut190*, Cut190**, in which the three C-terminal residues of Cut190* are deleted, and the inactive mutant, Cut190**S176A, in complex with metal ions. In addition to the previously observed closed, open and engaged forms, we determined the ejecting form, which would allow the product to irreversibly dissociate, followed by proceeding to the next cycle of reaction. These multiple forms would be stable or sub-stable states of Cut190, regulated by Ca2+ binding, and would be closely correlated with the enzyme function. Upon the deletion of the C-terminal residues, we found that the thermal stability increased while retaining the activity. The increased stability could be applied for the protein engineering of Cut190 for PET depolymerization as it requires the reaction above the glass transition temperature of PET.
Organizational Affiliation:
Graduate School of Life and Environmental Sciences, Kyoto Prefectural University, 1-5 Hangi-cho, Shimogamo, Sakyo-ku, Kyoto, Kyoto 606-8522, Japan.