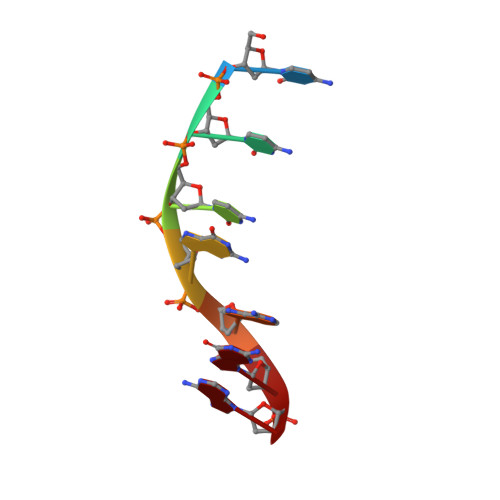
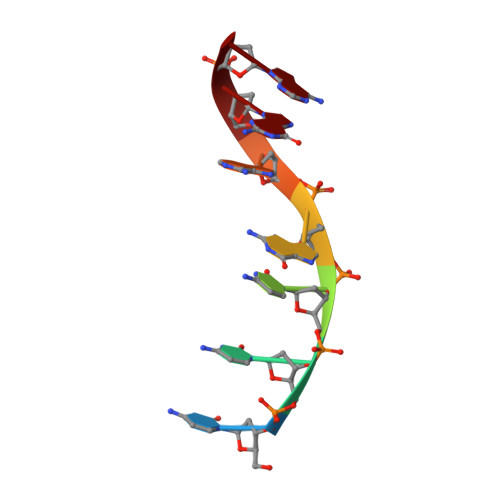
Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Novotny, A., Novotny, J., Kejnovska, I., Vorlickova, M., Fiala, R., Marek, R.(2021) Nucleic Acids Res 49: 11425-11437
- PubMed: 34718718
- DOI: https://doi.org/10.1093/nar/gkab915
- Primary Citation of Related Structures:
7BI0, 7BL0, 7BLM, 7BMA - PubMed Abstract:
Non-canonical forms of nucleic acids represent challenging objects for both structure-determination and investigation of their potential role in living systems. In this work, we uncover a structure adopted by GA repetition locked in a parallel homoduplex by an i-motif. A series of DNA oligonucleotides comprising GAGA segment and C3 clip is analyzed by NMR and CD spectroscopies to understand the sequence-structure-stability relationships. We demonstrate how the relative position of the homopurine GAGA segment and the C3 clip as well as single-base mutations (guanine deamination and cytosine methylation) affect base pairing arrangement of purines, i-motif topology and overall stability. We focus on oligonucleotides C3GAGA and methylated GAGAC3 exhibiting the highest stability and structural uniformity which allowed determination of high-resolution structures further analyzed by unbiased molecular dynamics simulation. We describe sequence-specific supramolecular interactions on the junction between homoduplex and i-motif blocks that contribute to the overall stability of the structures. The results show that the distinct structural motifs can not only coexist in the tight neighborhood within the same molecule but even mutually support their formation. Our findings are expected to have general validity and could serve as guides in future structure and stability investigations of nucleic acids.
Organizational Affiliation:
CEITEC - Central European Institute of Technology, Masaryk University, Kamenice 5, CZ-62500 Brno, Czechia.