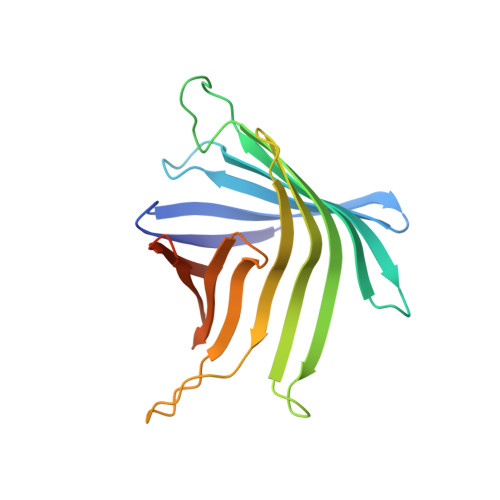
Structural basis of metabolite transport by the chloroplast outer envelope channel OEP21.
Gunsel, U., Klopfer, K., Hausler, E., Hitzenberger, M., Bolter, B., Sperl, L.E., Zacharias, M., Soll, J., Hagn, F.(2023) Nat Struct Mol Biol
- PubMed: 37156968
- DOI: https://doi.org/10.1038/s41594-023-00984-y
- Primary Citation of Related Structures:
7BGH - PubMed Abstract:
Triose phosphates (TPs) are the primary products of photosynthetic CO 2 fixation in chloroplasts, which need to be exported into the cytosol across the chloroplast inner envelope (IE) and outer envelope (OE) membranes to sustain plant growth. While transport across the IE is well understood, the mode of action of the transporters in the OE remains unclear. Here we present the high-resolution nuclear magnetic resonance (NMR) structure of the outer envelope protein 21 (OEP21) from garden pea, the main exit pore for TPs in C 3 plants. OEP21 is a cone-shaped β-barrel pore with a highly positively charged interior that enables binding and translocation of negatively charged metabolites in a competitive manner, up to a size of ~1 kDa. ATP stabilizes the channel and keeps it in an open state. Despite the broad substrate selectivity of OEP21, these results suggest that control of metabolite transport across the OE might be possible.
Organizational Affiliation:
Bavarian NMR Center (BNMRZ), Department of Bioscience, School of Natural Sciences, Technical University of Munich, Garching, Germany.