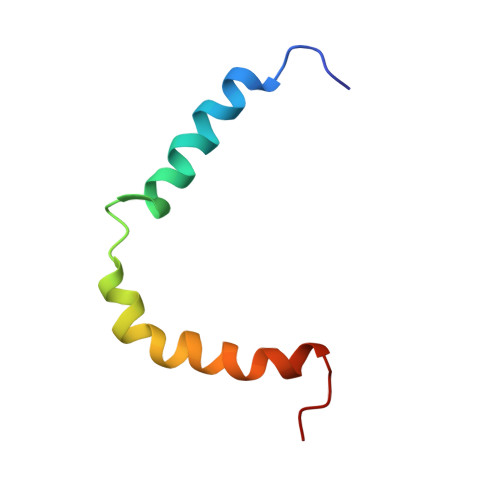
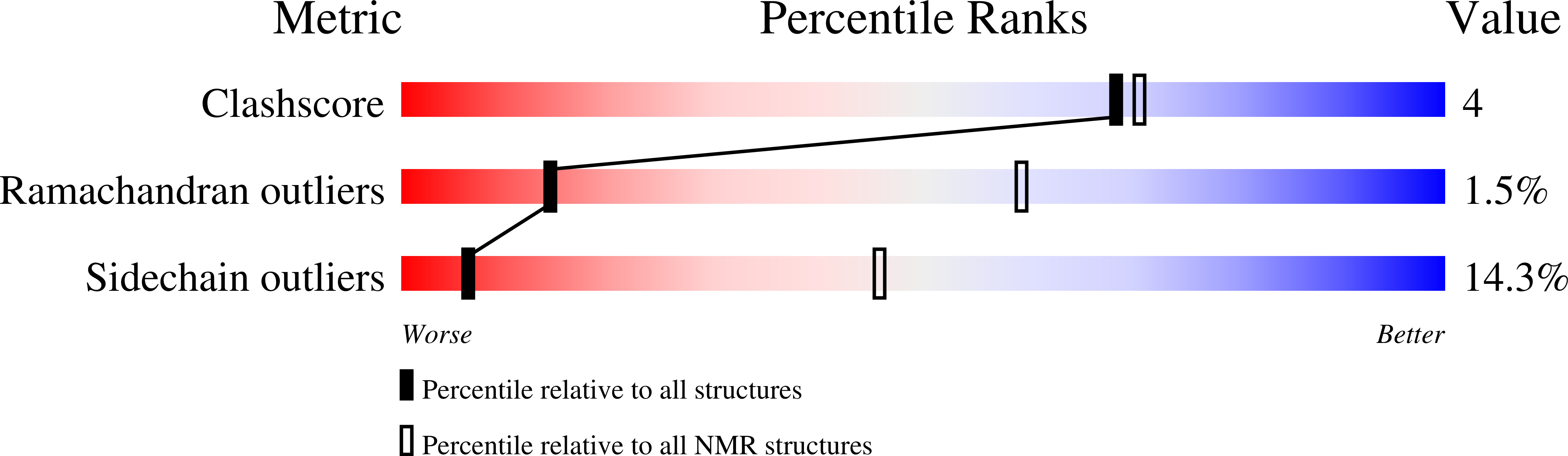Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR.
Pettersson, P., Ye, W., Jakob, M., Tannert, F., Klosgen, R.B., Maler, L.(2018) FEBS J 285: 1886-1906
- PubMed: 29654717
- DOI: https://doi.org/10.1111/febs.14452
- Primary Citation of Related Structures:
7B7O - PubMed Abstract:
The twin-arginine translocase (Tat) transports folded proteins across the cytoplasmic membrane of prokaryotes and the thylakoid membrane of plant chloroplasts. In Gram-negative bacteria and chloroplasts, the translocon consists of three subunits, TatA, TatB, and TatC, of which TatA is responsible for the actual membrane translocation of the substrate. Herein we report on the structure, dynamics, and lipid interactions of a fully functional C-terminally truncated 'core TatA' from Arabidopsis thaliana using solution-state NMR. Our results show that TatA consists of a short N-terminal transmembrane helix (TMH), a short connecting linker (hinge) and a long region with propensity to form an amphiphilic helix (APH). The dynamics of TatA were characterized using 15 N relaxation NMR in combination with model-free analysis. The TMH has order parameters characteristic of a well-structured helix, the hinge is somewhat less rigid, while the APH has lower order parameters indicating structural flexibility. The TMH is short with a surprisingly low protection from solvent, and only the first part of the APH is protected to some extent. In order to uncover possible differences in TatA's structure and dynamics in detergent compared to in a lipid bilayer, fast-tumbling bicelles and large unilamellar vesicles were used. Results indicate that the helicity of TatA increases in both the TMH and APH in the presence of lipids, and that the N-terminal part of the TMH is significantly more rigid. The results indicate that plant TatA has a significant structural plasticity and a capability to adapt to local environments.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Sweden.