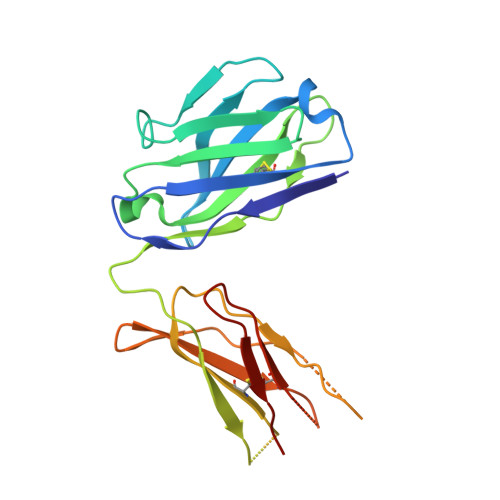
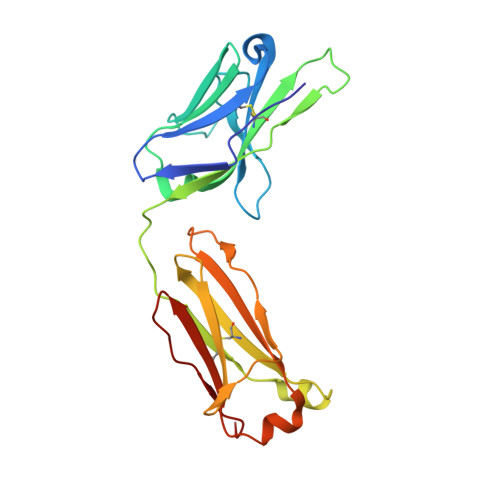
Higher-order immunoglobulin repertoire restrictions in CLL: the illustrative case of stereotyped subsets 2 and 169.
Gemenetzi, K., Psomopoulos, F., Carriles, A.A., Gounari, M., Minici, C., Plevova, K., Sutton, L.A., Tsagiopoulou, M., Baliakas, P., Pasentsis, K., Anagnostopoulos, A., Sandaltzopoulos, R., Rosenquist, R., Davi, F., Pospisilova, S., Ghia, P., Stamatopoulos, K., Degano, M., Chatzidimitriou, A.(2021) Blood 137: 1895-1904
- PubMed: 33036024
- DOI: https://doi.org/10.1182/blood.2020005216
- Primary Citation of Related Structures:
6ZTD - PubMed Abstract:
Chronic lymphocytic leukemia (CLL) major stereotyped subset 2 (IGHV3-21/IGLV3-21, ∼2.5% of all cases of CLL) is an aggressive disease variant, irrespective of the somatic hypermutation (SHM) status of the clonotypic IGHV gene. Minor stereotyped subset 169 (IGHV3-48/IGLV3-21, ∼0.2% of all cases of CLL) is related to subset 2, as it displays a highly similar variable antigen-binding site. We further explored this relationship through next-generation sequencing and crystallographic analysis of the clonotypic B-cell receptor immunoglobulin. Branching evolution of the predominant clonotype through intraclonal diversification in the context of ongoing SHM was evident in both heavy and light chain genes of both subsets. Molecular similarities between the 2 subsets were highlighted by the finding of shared SHMs within both the heavy and light chain genes in all analyzed cases at either the clonal or subclonal level. Particularly noteworthy in this respect was a ubiquitous SHM at the linker region between the variable and the constant domain of the IGLV3-21 light chains, previously reported as critical for immunoglobulin homotypic interactions underlying cell-autonomous signaling capacity. Notably, crystallographic analysis revealed that the IGLV3-21-bearing CLL subset 169 immunoglobulin retains the same geometry and contact residues for the homotypic intermolecular interaction observed in subset 2, including the SHM at the linker region, and, from a molecular standpoint, belong to a common structural mode of autologous recognition. Collectively, our findings document that stereotyped subsets 2 and 169 are very closely related, displaying shared immunoglobulin features that can be explained only in the context of shared functional selection.
Organizational Affiliation:
Institute of Applied Biosciences, Centre for Research and Technology Hellas, Thessaloniki, Greece.