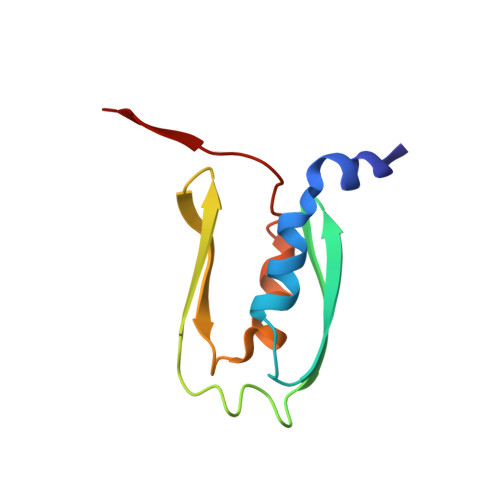
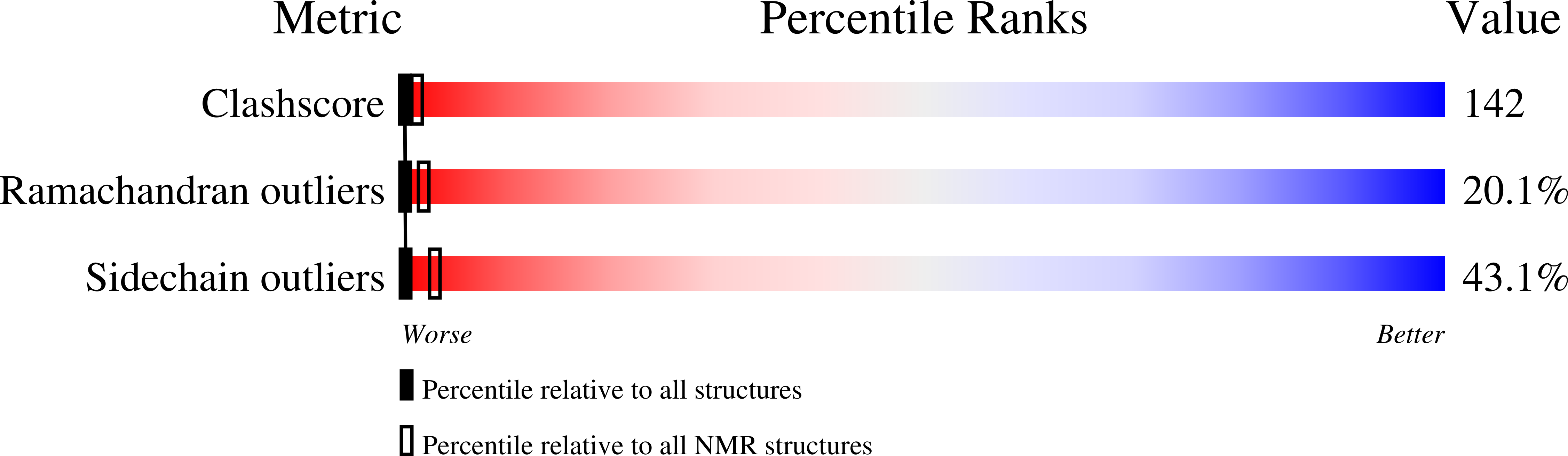Protein yoga: Conformational versatility of the Hemolysin II C-terminal domain detailed by NMR structures for multiple states.
Kaplan, A.R., Olson, R., Alexandrescu, A.T.(2021) Protein Sci 30: 990-1005
- PubMed: 33733504
- DOI: https://doi.org/10.1002/pro.4066
- Primary Citation of Related Structures:
6WA1 - PubMed Abstract:
The C-terminal domain of Bacillus cereus hemolysin II (HlyIIC), stabilizes the trans-membrane-pore formed by the HlyII toxin and may aid in target cell recognition. Initial efforts to determine the NMR structure of HlyIIC were hampered by cis/trans isomerization about the single proline at position 405 that leads to doubling of NMR resonances. We used the mutant P405M-HlyIIC that eliminates the cis proline to determine the NMR structure of the domain, which revealed a novel fold. Here, we extend earlier studies to the NMR structure determination of the cis and trans states of WT-HlyIIC that exist simultaneously in solution. The primary structural differences between the cis and trans states are in the loop that contains P405, and structurally adjacent loops. Thermodynamic linkage analysis shows that at 25 C the cis proline, which already has a large fraction of 20% in the unfolded protein, increases to 50% in the folded state due to coupling with the global stability of the domain. The P405M or P405A substitutions eliminate heterogeneity due to proline isomerization but lead to the formation of a new dimeric species. The NMR structure of the dimer shows that it is formed through domain-swapping of strand β5, the last segment of secondary structure following P405. The presence of P405 in WT-HlyIIC strongly disfavors the dimer compared to the P405M-HlyIIC or P405A-HlyIIC mutants. The WT proline may thus act as a "gatekeeper," warding off aggregative misfolding.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of Connecticut, Storrs, Connecticut, USA.