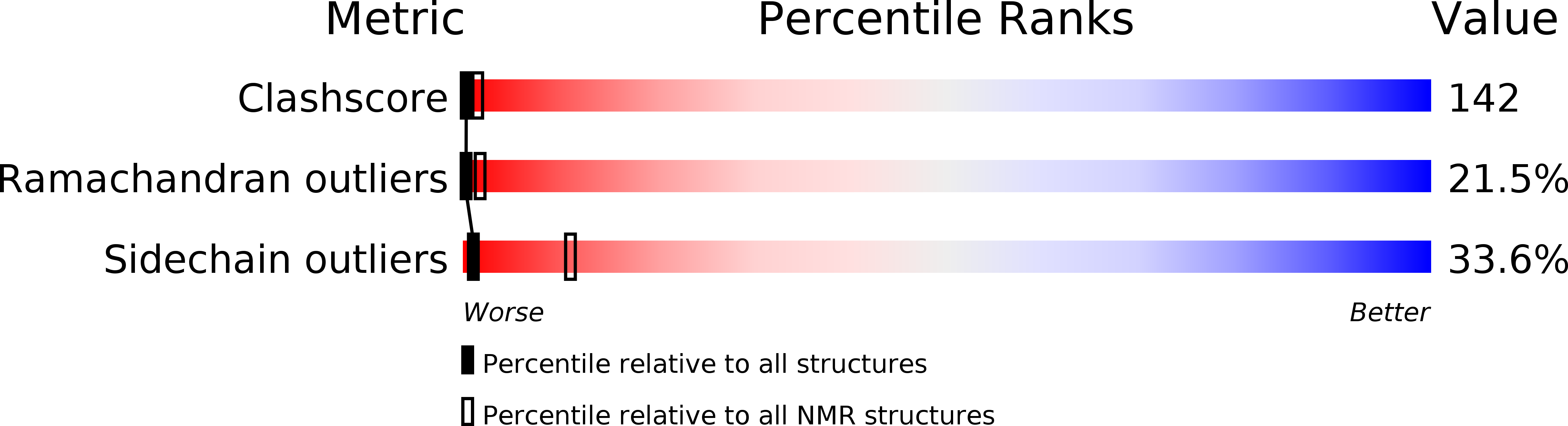Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Yang, Y., Lin, J., Harrington, A., Cornilescu, G., Lau, G.W., Tal-Gan, Y.(2020) Proc Natl Acad Sci U S A 117: 1689-1699
- PubMed: 31915298
- DOI: https://doi.org/10.1073/pnas.1915812117
- Primary Citation of Related Structures:
6OBW, 6OC2, 6OC4, 6OLD, 6V1N - PubMed Abstract:
Streptococcus pneumoniae is an opportunistic human pathogen that utilizes the competence regulon, a quorum-sensing circuitry, to acquire antibiotic resistance genes and initiate its attack on the human host. Interception of the competence regulon can therefore be utilized to study S. pneumoniae cell-cell communication and behavioral changes, as well as attenuate S. pneumoniae infectivity. Herein we report the design and synthesis of cyclic dominant negative competence-stimulating peptide (dnCSP) analogs capable of intercepting the competence regulon in both S. pneumoniae specificity groups with activities at the low nanomolar range. Structural analysis of lead analogs provided important insights as to the molecular mechanism that drives CSP receptor binding and revealed that the pan-group cyclic CSPs exhibit a chimeric hydrophobic patch conformation that resembles the hydrophobic patches required for both ComD1 and ComD2 binding. Moreover, the lead cyclic dnCSP, CSP1-E1A-cyc(Dap6E10), was found to possess superior pharmacological properties, including improved resistance to enzymatic degradation, while remaining nontoxic. Lastly, CSP1-E1A-cyc(Dap6E10) was capable of attenuating mouse mortality during acute pneumonia caused by both group 1 and group 2 S. pneumoniae strains. This cyclic pan-group dnCSP is therefore a promising drug lead scaffold against S. pneumoniae infections that could be administered individually or utilized in combination therapy to augment the effects of current antimicrobial agents.
Organizational Affiliation:
Department of Chemistry, University of Nevada, Reno, Reno, NV 89557.