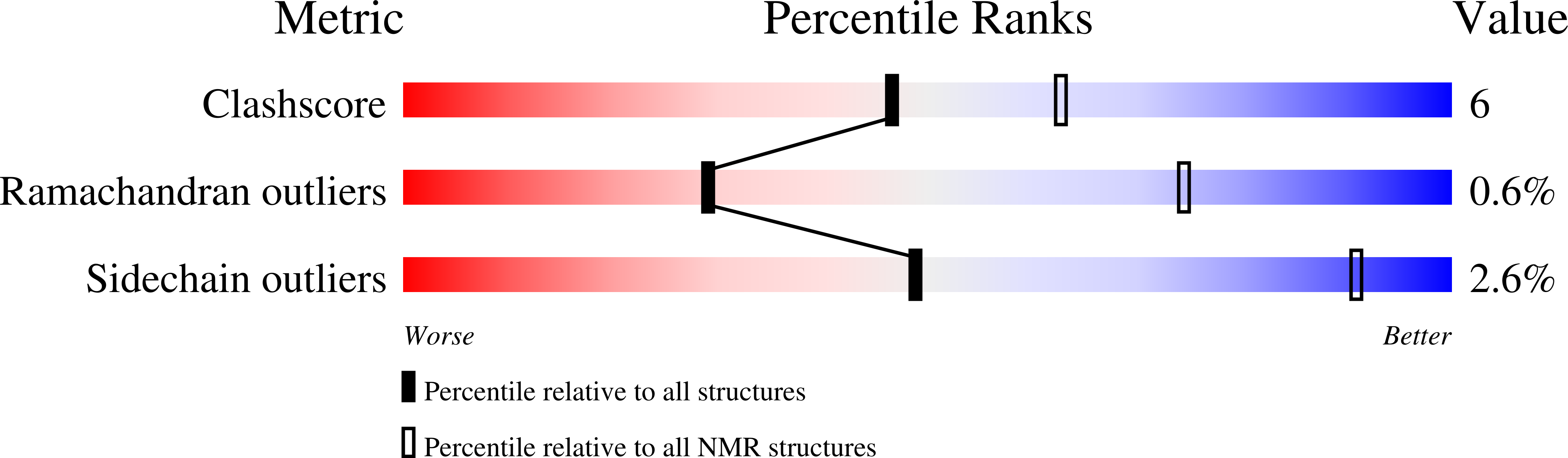Solution Structure of Tubuliform Spidroin N-Terminal Domain and Implications for pH Dependent Dimerization.
Sede, M., Fridmanis, J., Otikovs, M., Johansson, J., Rising, A., Kronqvist, N., Jaudzems, K.(2022) Front Mol Biosci 9: 936887-936887
- PubMed: 35775078
- DOI: https://doi.org/10.3389/fmolb.2022.936887
- Primary Citation of Related Structures:
6TV5 - PubMed Abstract:
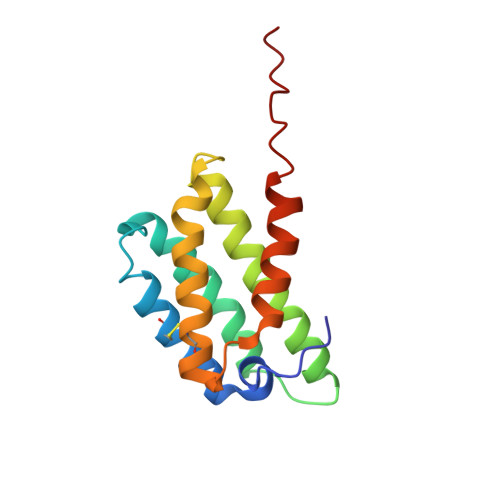
The spidroin N-terminal domain (NT) is responsible for high solubility and pH-dependent assembly of spider silk proteins during storage and fiber formation, respectively. It forms a monomeric five-helix bundle at neutral pH and dimerizes at lowered pH, thereby firmly interconnecting the spidroins. Mechanistic studies with the NTs from major ampullate, minor ampullate, and flagelliform spidroins (MaSp, MiSp, and FlSp) have shown that the pH dependency is conserved between different silk types, although the residues that mediate this process can differ. Here we study the tubuliform spidroin (TuSp) NT from Argiope argentata , which lacks several well conserved residues involved in the dimerization of other NTs. We solve its structure at low pH revealing an antiparallel dimer of two five-α-helix bundles, which contrasts with a previously determined Nephila antipodiana TuSp NT monomer structure. Further, we study a set of mutants and find that the residues participating in the protonation events during dimerization are different from MaSp and MiSp NT. Charge reversal of one of these residues (R117 in TuSp) results in significantly altered electrostatic interactions between monomer subunits. Altogether, the structure and mutant studies suggest that TuSp NT monomers assemble by elimination of intramolecular repulsive charge interactions, which could lead to slight tilting of α-helices.
Organizational Affiliation:
Department of Physical Organic Chemistry, Latvian Institute of Organic Synthesis, Riga, Latvia.