NMR structure of Pseudomonas fluorescens CopC
Persson, K.C., Mayzel, M., Karlsson, B.G., Peciulyte, A., Olsson, L., Wittung Stafshede, P., Salomon Johansen, K., Horvath, I.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative copper resistance protein | 97 | Pseudomonas fluorescens | Mutation(s): 0 Gene Names: PFLU_3946 | 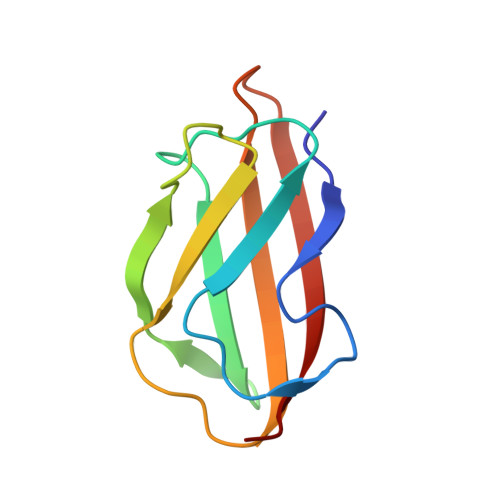 | |
UniProt | |||||
Find proteins for C3JYL7 (Pseudomonas fluorescens (strain SBW25)) Explore C3JYL7 Go to UniProtKB: C3JYL7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C3JYL7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Commission | Denmark | 608473 |