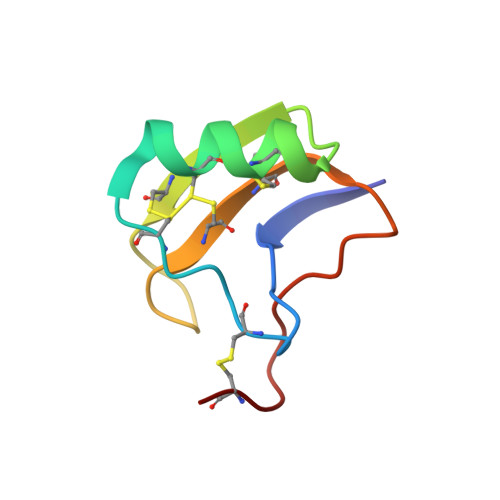
Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Mineev, K.S., Kuzmenkov, A.I., Arseniev, A.S., Vassilevski, A.A.(2021) Proteins
- PubMed: 33713480
- DOI: https://doi.org/10.1002/prot.26074
- Primary Citation of Related Structures:
6THI - PubMed Abstract:
Old world scorpions produce an abundance of toxins called α-NaTx, which interfere with the fast inactivation of voltage-gated sodium channels. Their selectivity to channels of mammals or insects depends on a part of toxin named the specificity module. We report here the spatial structure of a major and broadly active toxin MeuNaTxα-1 from the venom of Mesobuthus eupeus. Notably, its specificity module is markedly different from other α-NaTx with known 3D structure. Close inspection shows that its conformation is a result of an interplay between protein motifs such as the nest and niche, which eventually shape α-NaTx structural diversity.
- Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russian Federation.
Organizational Affiliation: