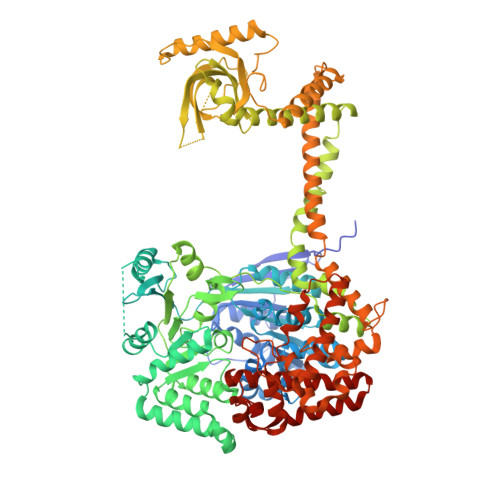
The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Lingaraju, M., Johnsen, D., Schlundt, A., Langer, L.M., Basquin, J., Sattler, M., Heick Jensen, T., Falk, S., Conti, E.(2019) Nat Commun 10: 3393-3393
- PubMed: 31358741
- DOI: https://doi.org/10.1038/s41467-019-11339-x
- Primary Citation of Related Structures:
6RO1 - PubMed Abstract:
The nuclear exosome and its essential co-factor, the RNA helicase MTR4, play crucial roles in several RNA degradation pathways. Besides unwinding RNA substrates for exosome-mediated degradation, MTR4 associates with RNA-binding proteins that function as adaptors in different RNA processing and decay pathways. Here, we identify and characterize the interactions of human MTR4 with a ribosome processing adaptor, NVL, and with ZCCHC8, an adaptor involved in the decay of small nuclear RNAs. We show that the unstructured regions of NVL and ZCCHC8 contain short linear motifs that bind the MTR4 arch domain in a mutually exclusive manner. These short sequences diverged from the arch-interacting motif (AIM) of yeast rRNA processing factors. Our results suggest that nuclear exosome adaptors have evolved canonical and non-canonical AIM sequences to target human MTR4 and demonstrate the versatility and specificity with which the MTR4 arch domain can recruit a repertoire of different RNA-binding proteins.
Organizational Affiliation:
Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, D-82152, Martinsried, Germany.