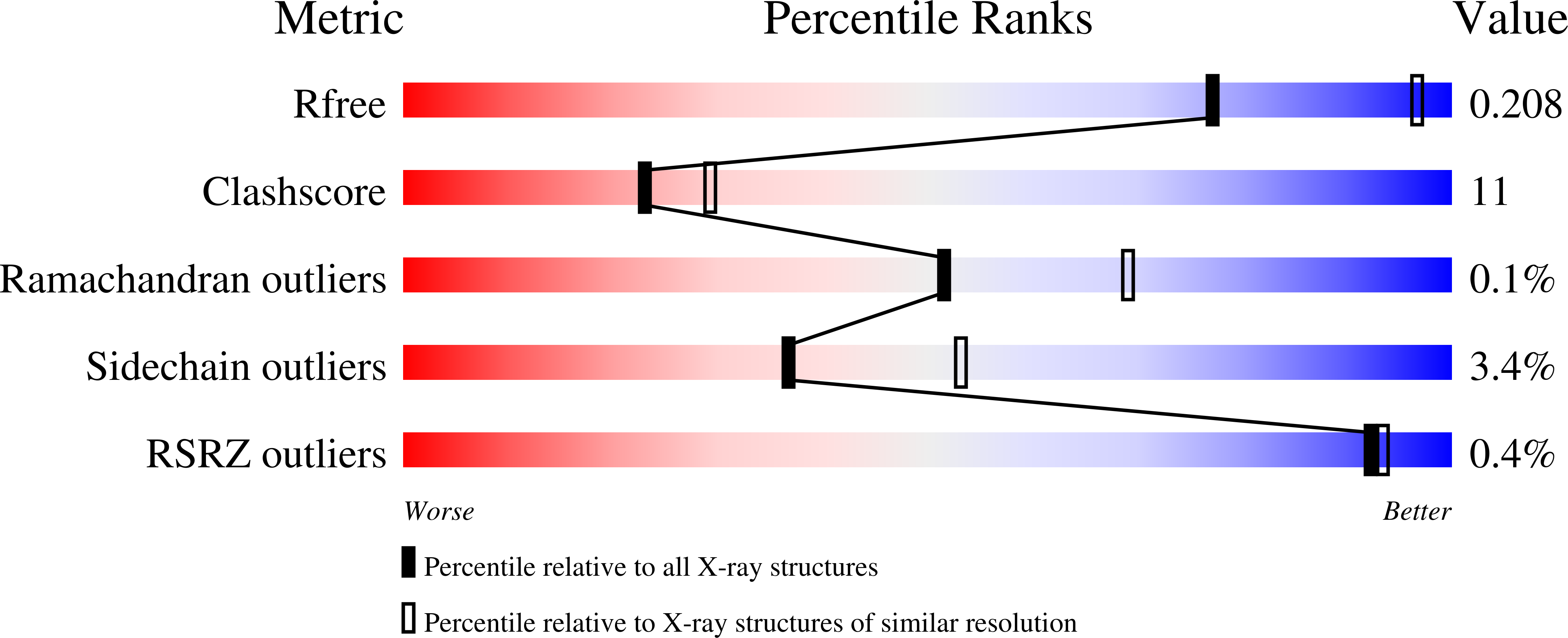
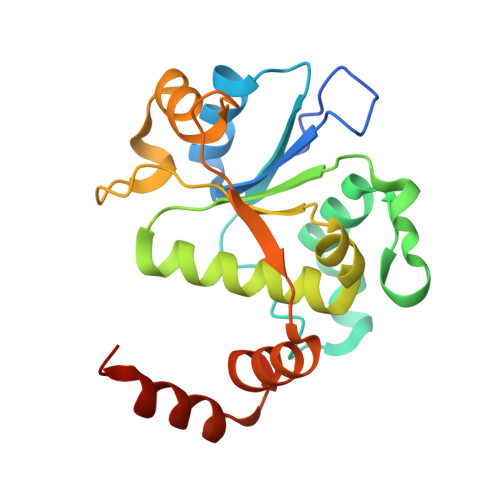
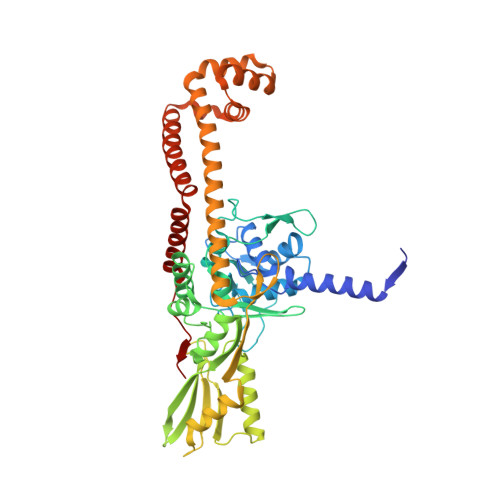
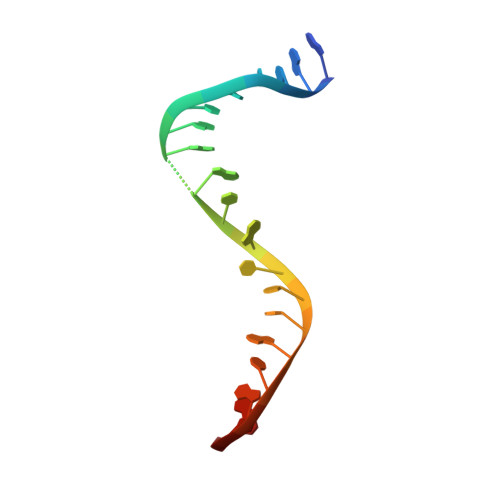
Structure-guided design of antibacterials that allosterically inhibit DNA gyrase.
Thalji, R.K., Raha, K., Andreotti, D., Checchia, A., Cui, H., Meneghelli, G., Profeta, R., Tonelli, F., Tommasi, S., Bakshi, T., Donovan, B.T., Howells, A., Jain, S., Nixon, C., Quinque, G., McCloskey, L., Bax, B.D., Neu, M., Chan, P.F., Stavenger, R.A.(2019) Bioorg Med Chem Lett 29: 1407-1412
- PubMed: 30962087
- DOI: https://doi.org/10.1016/j.bmcl.2019.03.029
- Primary Citation of Related Structures:
6QX1, 6QX2 - PubMed Abstract:
A series of DNA gyrase inhibitors were designed based on the X-ray structure of a parent thiophene scaffold with the objective to improve biochemical and whole-cell antibacterial activity, while reducing cardiac ion channel activity. The binding mode and overall design hypothesis of one series was confirmed with a co-crystal structure with DNA gyrase. Although some analogs retained both biochemical activity and whole-cell antibacterial activity, we were unable to significantly improve the activity of the series and analogs retained activity against the cardiac ion channels, therefore we stopped optimization efforts.
Organizational Affiliation:
GlaxoSmithKline, Collegeville, PA 19426, USA.