Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA
Halabelian, L., Zeng, H., Li, Y., Bountra, C., Edwards, A.M., Arrowsmith, C.H., Structural Genomics Consortium (SGC)To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein | 276 | Homo sapiens | Mutation(s): 0 Gene Names: HMCES, C3orf37, DC12, SRAPD1 EC: 3.4 | 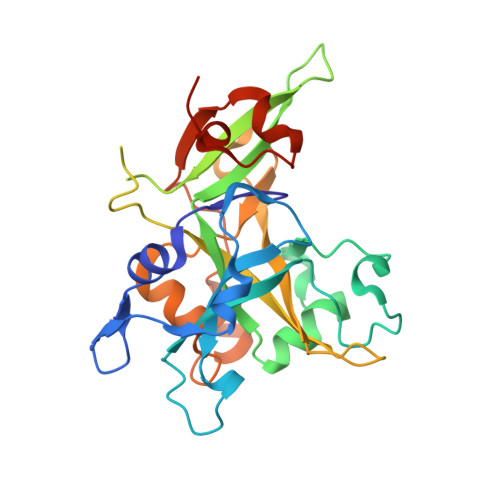 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96FZ2 (Homo sapiens) Explore Q96FZ2 Go to UniProtKB: Q96FZ2 | |||||
PHAROS: Q96FZ2 GTEx: ENSG00000183624 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96FZ2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*AP*AP*CP*GP*TP*TP*GP*TP*TP*TP*TP*T)-3') | 13 | synthetic construct | 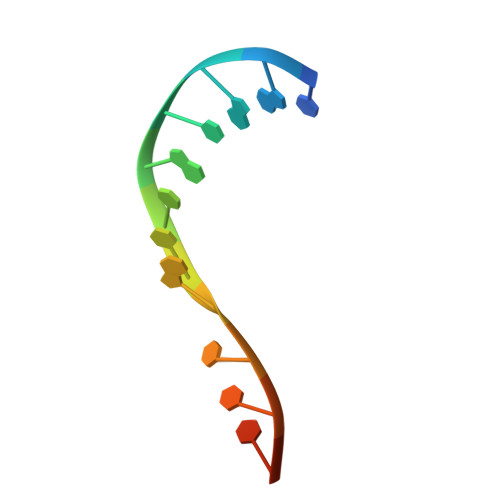 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| UNX Query on UNX | AA [auth A] BA [auth A] CA [auth A] DA [auth A] E [auth A] | UNKNOWN ATOM OR ION X |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.729 | α = 90 |
| b = 139.828 | β = 99.99 |
| c = 66.613 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| REFMAC | phasing |