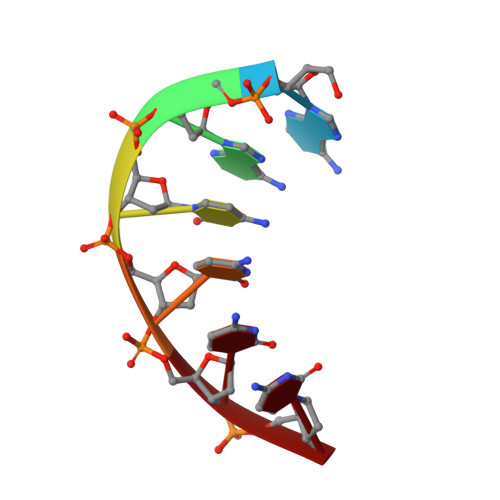
Atomic Structure of a Fluorescent Ag8Cluster Templated by a Multistranded DNA Scaffold.
Huard, D.J.E., Demissie, A., Kim, D., Lewis, D., Dickson, R.M., Petty, J.T., Lieberman, R.L.(2019) J Am Chem Soc 141: 11465-11470
- PubMed: 30562465
- DOI: https://doi.org/10.1021/jacs.8b12203
- Primary Citation of Related Structures:
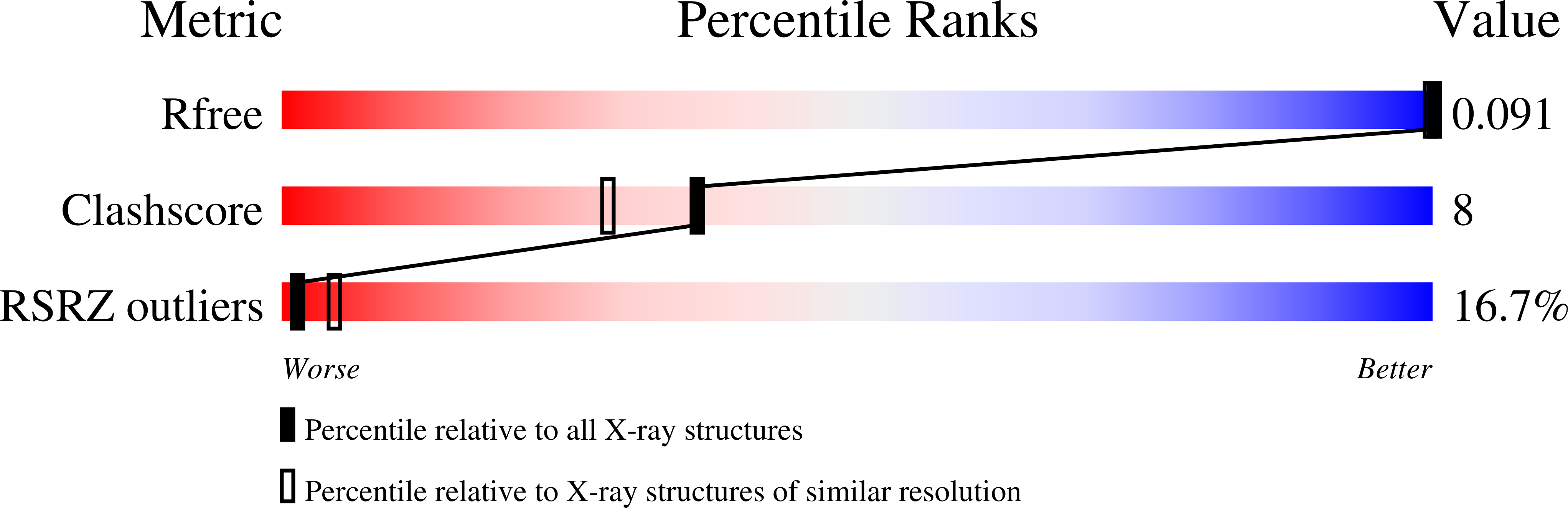
6NIZ - PubMed Abstract:
Multinuclear silver clusters encapsulated by DNA exhibit size-tunable emission spectra and rich photophysics, but their atomic organization is poorly understood. Herein, we describe the structure of one such hybrid chromophore, a green-emitting Ag 8 cluster arranged in a Big Dipper-shape bound to the oligonucleotide A 2 C 4 . Three 3' cytosine metallo-base pairs stabilize a parallel A-form-like duplex with a 5' adenine-rich pocket, which binds a metallic, trapezoidal-shaped Ag 5 moiety via Ag-N bonds to endo- and exocyclic nitrogens of cytosine and adenine. The unique DNA configuration, constrained coordination environment, and templated Ag 8 cluster arrangement highlight the reciprocity between the silvers and DNA in adopting this structure. These first atomic details of a DNA-encapsulated Ag cluster fluorophore illuminate many aspects of biological assembly, nanoscience, and metal cluster photophysics.
Organizational Affiliation:
School of Chemistry & Biochemistry and Parker H. Petit Institute for Bioengineering and Bioscience , Georgia Institute of Technology , Atlanta , Georgia 30332 , United States.