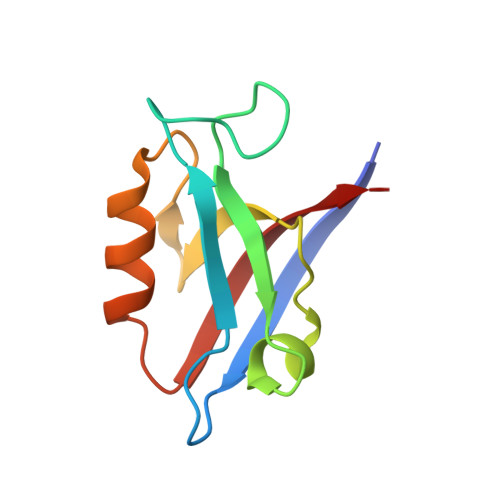
SGEF forms a complex with Scribble and Dlg1 and regulates epithelial junctions and contractility.
Awadia, S., Huq, F., Arnold, T.R., Goicoechea, S.M., Sun, Y.J., Hou, T., Kreider-Letterman, G., Massimi, P., Banks, L., Fuentes, E.J., Miller, A.L., Garcia-Mata, R.(2019) J Cell Biol 218: 2699-2725
- PubMed: 31248911
- DOI: https://doi.org/10.1083/jcb.201811114
- Primary Citation of Related Structures:
6MYE, 6MYF - PubMed Abstract:
The canonical Scribble polarity complex is implicated in regulation of epithelial junctions and apical polarity. Here, we show that SGEF, a RhoG-specific GEF, forms a ternary complex with Scribble and Dlg1, two members of the Scribble complex. SGEF targets to apical junctions in a Scribble-dependent fashion and functions in the regulation of actomyosin-based contractility and barrier function at tight junctions as well as E-cadherin-mediated formation of adherens junctions. Surprisingly, SGEF does not control the establishment of polarity. However, in 3D cysts, SGEF regulates the formation of a single open lumen. Interestingly, SGEF's nucleotide exchange activity regulates the formation and maintenance of adherens junctions, and in cysts the number of lumens formed, whereas SGEF's scaffolding activity is critical for regulation of actomyosin contractility and lumen opening. We propose that SGEF plays a key role in coordinating junctional assembly and actomyosin contractility by bringing together Scribble and Dlg1 and targeting RhoG activation to cell-cell junctions.
Organizational Affiliation:
Department of Biological Sciences, The University of Toledo, Toledo, OH.