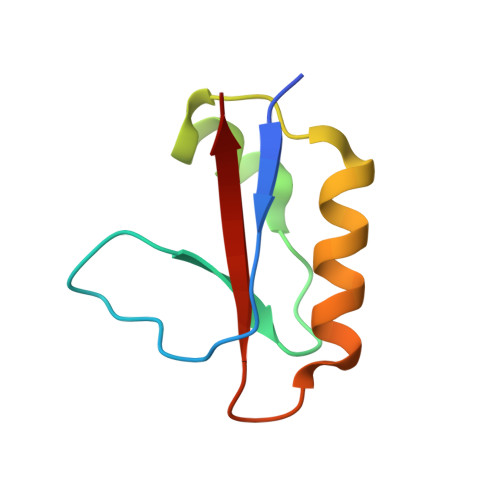
Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Kim, I., Koo, J., An, S.Y., Hong, S., Ka, D., Kim, E.H., Bae, E., Suh, J.Y.(2020) Nucleic Acids Res 48: 9959-9968
- PubMed: 32810226
- DOI: https://doi.org/10.1093/nar/gkaa690
- Primary Citation of Related Structures:
6M3N - PubMed Abstract:
The CRISPR-Cas system provides adaptive immunity for bacteria and archaea to combat invading phages and plasmids. Phages evolved anti-CRISPR (Acr) proteins to neutralize the host CRISPR-Cas immune system as a counter-defense mechanism. AcrIF7 in Pseudomonas aeruginosa prophages strongly inhibits the type I-F CRISPR-Cas system. Here, we determined the solution structure of AcrIF7 and identified its target, Cas8f of the Csy complex. AcrIF7 adopts a novel β1β2α1α2β3 fold and interacts with the target DNA binding site of Cas8f. Notably, AcrIF7 competes with AcrIF2 for the same binding interface on Cas8f without common structural motifs. AcrIF7 binding to Cas8f is driven mainly by electrostatic interactions that require position-specific surface charges. Our findings suggest that Acrs of divergent origin may have acquired specificity to a common target through convergent evolution of their surface charge configurations.
Organizational Affiliation:
Department of Agricultural Biotechnology, Seoul National University, Seoul 08826, Korea.