Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Min, C.A., Yun, J.S., Choi, E.H., Hwang, U.W., Cho, D.H., Yoon, J.H., Chang, J.H.(2019) Crystals (Basel) 9
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2019) Crystals (Basel) 9
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fatty acid amide hydrolase | 571 | Candida albicans | Mutation(s): 0 Gene Names: CaJ7.0332, CaO19.5169 | 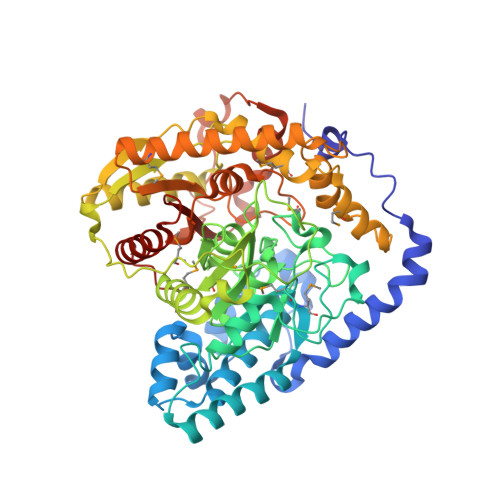 | |
UniProt | |||||
Find proteins for G1UA68 (Candida albicans) Explore G1UA68 Go to UniProtKB: G1UA68 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G1UA68 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 84.874 | α = 90 |
| b = 68.753 | β = 99.628 |
| c = 100.979 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (MoE, Korea) | Korea, Republic Of | Bokhyeon Research Fund 2017 |