Structural and functional analyses of the lipase CinB from Enterobacter asburiae.
Shang, F., Lan, J., Liu, W., Chen, Y., Wang, L., Zhao, J., Chen, J., Gao, P., Ha, N.C., Quan, C., Nam, K.H., Xu, Y.(2019) Biochem Biophys Res Commun 519: 274-279
- PubMed: 31493870
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.166
- Primary Citation of Related Structures:
6KMO - PubMed Abstract:
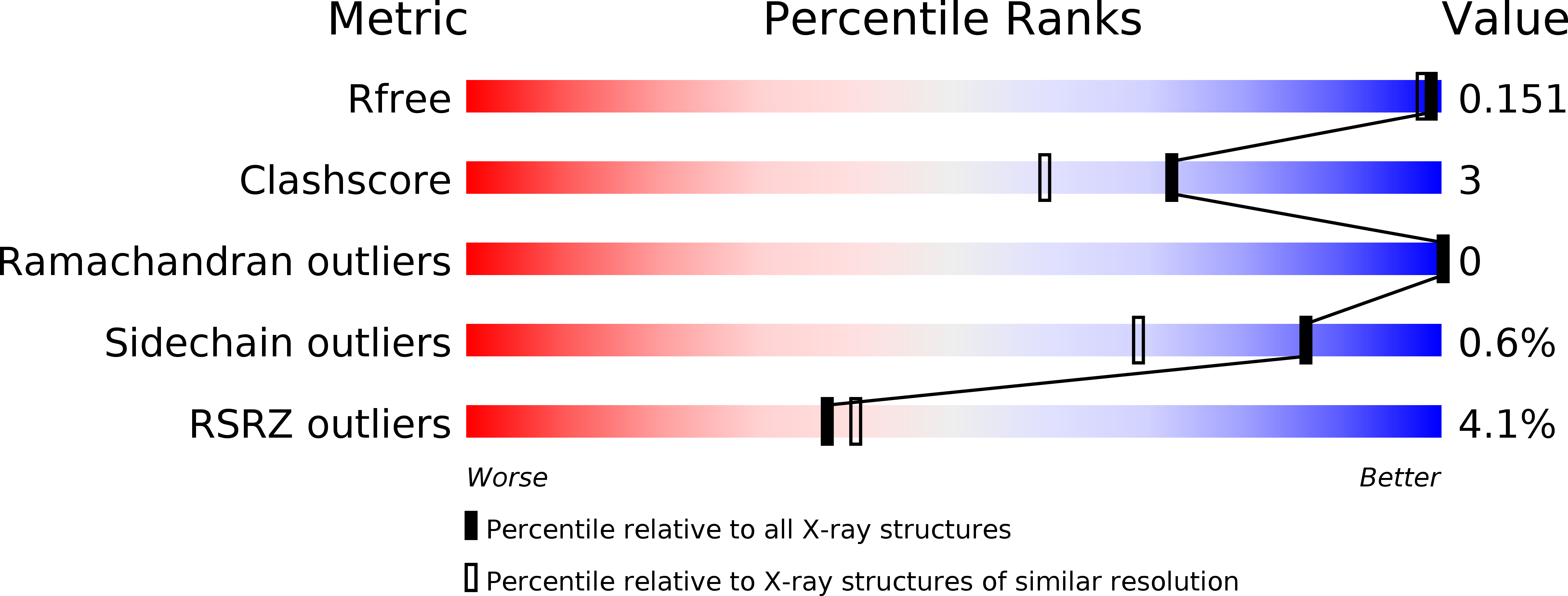
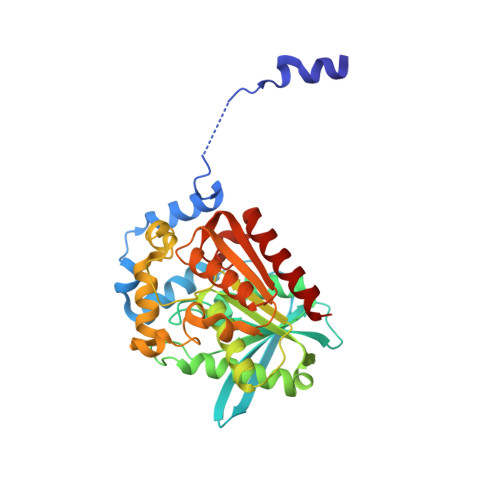
Lipases are widely present in various plants, animals and microorganisms, constituting a large category of enzymes. They have the ability to catalyze the cleavage of ester bonds. The lipase CinB from Enterobacter asburiae (E. asburiae) is an acetyl esterase. The primary amino acid sequence suggests that the EaCinB protein belongs to the α/β-hydrolase (ABH) superfamily of the esterase/lipase superfamily. However, its molecular functions have not yet been determined. Here, we report the crystal structure of E. asburiae CinB at a 1.45 Å resolution. EaCinB contains a signal peptide, cap domain and catalytic domain. The active site of EaCinB contains the catalytic triad (Ser180-His307-Asp277) on the catalytic domain. The oxyanion hole is composed of Gly106 and Gly107 within the conserved sequence motif HGGG (amino acid residues 106-109). The substrate is accessible between the α1 and α2 helices or the α1 helix and catalytic domain. Narrow substrate pockets are formed by the α2 helix of the cap domain. Site-directed mutagenesis showed that EaCinB-W208H exhibits a higher catalytic ability than EaCinB-WT by approximately nine times. Our results provide insight into the molecular function of EaCinB.
Organizational Affiliation:
Department of Bioengineering, College of Life Science, Dalian Minzu University, Dalian, 116600, Liaoning, China; Key Laboratory of Biotechnology and Bioresources Utilization (Dalian Minzu University), Ministry of Education, China.