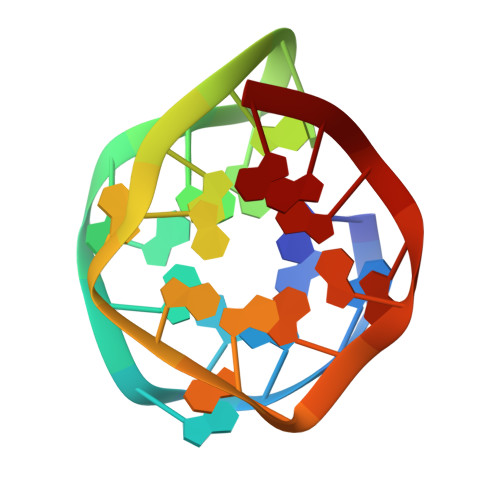
Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Mashima, T., Lee, J.H., Kamatari, Y.O., Hayashi, T., Nagata, T., Nishikawa, F., Nishikawa, S., Kinoshita, M., Kuwata, K., Katahira, M.(2020) Sci Rep 10: 4934-4934
- PubMed: 32188933
- DOI: https://doi.org/10.1038/s41598-020-61966-4
- Primary Citation of Related Structures:
6K84 - PubMed Abstract:
Prion diseases comprise a fatal neuropathy caused by the conversion of prion protein from a cellular (PrP C ) to a pathological (PrP Sc ) isoform. Previously, we obtained an RNA aptamer, r(GGAGGAGGAGGA) (R12), that folds into a unique G-quadruplex. The R12 homodimer binds to a PrP C molecule, inhibiting PrP C -to-PrP Sc conversion. Here, we developed a new RNA aptamer, r(GGAGGAGGAGGAGGAGGAGGAGGA) (R24), where two R12s are tandemly connected. The 50% inhibitory concentration for the formation of PrP Sc (IC 50 ) of R24 in scrapie-infected cell lines was ca. 100 nM, i.e., much lower than that of R12 by two orders. Except for some antibodies, R24 exhibited the lowest recorded IC 50 and the highest anti-prion activity. We also developed a related aptamer, r(GGAGGAGGAGGA-A-GGAGGAGGAGGA) (R12-A-R12), IC 50 being ca. 500 nM. The structure of a single R12-A-R12 molecule determined by NMR resembled that of the R12 homodimer. The quadruplex structure of either R24 or R12-A-R12 is unimolecular, and therefore the structure could be stably formed when they are administered to a prion-infected cell culture. This may be the reason they can exert high anti-prion activity.
Organizational Affiliation:
Institute of Advanced Energy, Kyoto University, Kyoto, 611-0011, Japan.