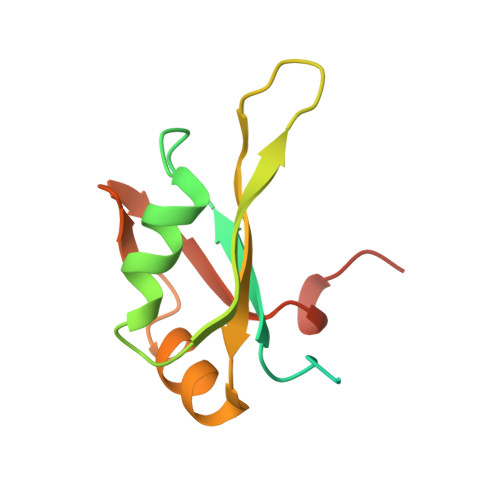
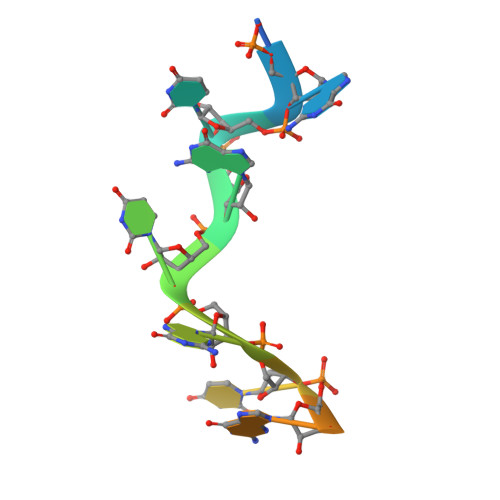
Structural basis for mRNA recognition by human RBM38.
Qian, K., Li, M., Wang, J., Zhang, M., Wang, M.(2020) Biochem J 477: 161-172
- PubMed: 31860021
- DOI: https://doi.org/10.1042/BCJ20190652
- Primary Citation of Related Structures:
6JVX, 6JVY - PubMed Abstract:
RNA-binding protein RBM38 was reported to bind the mRNA of several p53-related genes through its RRM domain and to up-regulate or down-regulate protein translation by increasing mRNA stability or recruitment of other effector proteins. The recognition mechanism, however, for RNA-binding of RBM38 remains unclear. Here, we report the crystal structure of the RRM domain of human RBM38 in complex with a single-stranded RNA. Our structural and biological results revealed that RBM38 recognizes G(U/C/A)GUG sequence single-stranded RNA in a sequence-specific and structure-specific manner. Two phenylalanine stacked with bases of RNA were crucial for RNA binding, and a series of hydrogen bonds between the base atoms of RNA and main-chain or side-chain atoms of RBM38 determine the sequence-specific recognition. Our results revealed the RNA-recognition mechanism of human RBM38 and provided structural information for understanding the RNA-binding property of RBM38.
Organizational Affiliation:
School of Life Sciences, Anhui University, Hefei, Anhui 230601, China.