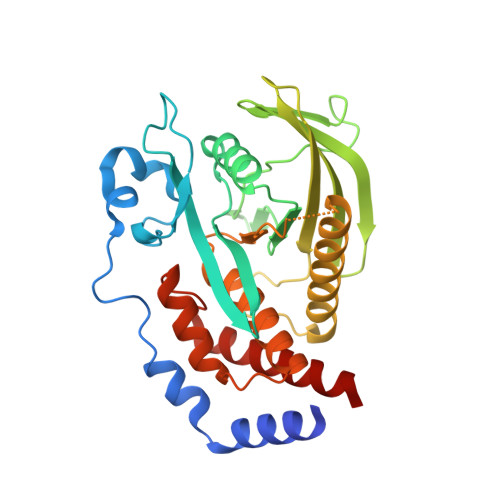
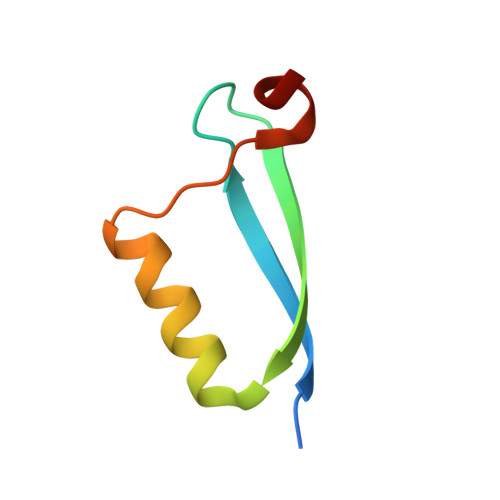
Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus.
Yun, H.Y., Kim, M.W., Lee, H.S., Kim, W., Shin, J.H., Kim, H., Shin, H.C., Park, H., Oh, B.H., Kim, W.K., Bae, K.H., Lee, S.C., Lee, E.W., Ku, B., Kim, S.J.(2019) PLoS Biol 17: e3000367-e3000367
- PubMed: 31323018
- DOI: https://doi.org/10.1371/journal.pbio.3000367
- Primary Citation of Related Structures:
6IWD - PubMed Abstract:
Human papillomaviruses (HPVs) are causative agents of various diseases associated with cellular hyperproliferation, including cervical cancer, one of the most prevalent tumors in women. E7 is one of the two HPV-encoded oncoproteins and directs recruitment and subsequent degradation of tumor-suppressive proteins such as retinoblastoma protein (pRb) via its LxCxE motif. E7 also triggers tumorigenesis in a pRb-independent pathway through its C-terminal domain, which has yet been largely undetermined, with a lack of structural information in a complex form with a host protein. Herein, we present the crystal structure of the E7 C-terminal domain of HPV18 belonging to the high-risk HPV genotypes bound to the catalytic domain of human nonreceptor-type protein tyrosine phosphatase 14 (PTPN14). They interact directly and potently with each other, with a dissociation constant of 18.2 nM. Ensuing structural analysis revealed the molecular basis of the PTPN14-binding specificity of E7 over other protein tyrosine phosphatases and also led to the identification of PTPN21 as a direct interacting partner of E7. Disruption of HPV18 E7 binding to PTPN14 by structure-based mutagenesis impaired E7's ability to promote keratinocyte proliferation and migration. Likewise, E7 binding-defective PTPN14 was resistant for degradation via proteasome, and it was much more effective than wild-type PTPN14 in attenuating the activity of downstream effectors of Hippo signaling and negatively regulating cell proliferation, migration, and invasion when examined in HPV18-positive HeLa cells. These results therefore demonstrated the significance and therapeutic potential of the intermolecular interaction between HPV E7 and host PTPN14 in HPV-mediated cell transformation and tumorigenesis.
Organizational Affiliation:
Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea.