(1)H, (13)C, and (15)N resonance assignments for the pro-inflammatory cytokine interleukin-36alpha.
Goradia, N., Wissbrock, A., Wiedemann, C., Bordusa, F., Ramachandran, R., Imhof, D., Ohlenschlaeger, O.(2016) Biomol NMR Assign 10: 329-333
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2016) Biomol NMR Assign 10: 329-333
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Interleukin-36 alpha | 158 | Homo sapiens | Mutation(s): 0 Gene Names: IL36A, FIL1E, IL1E, IL1F6 | 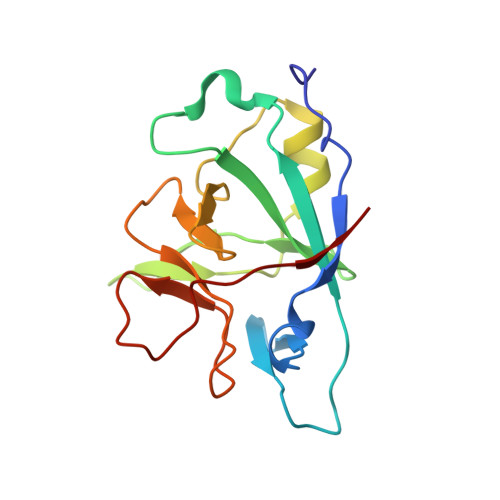 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UHA7 (Homo sapiens) Explore Q9UHA7 Go to UniProtKB: Q9UHA7 | |||||
PHAROS: Q9UHA7 GTEx: ENSG00000136694 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UHA7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation | Germany | FOR 1738 |