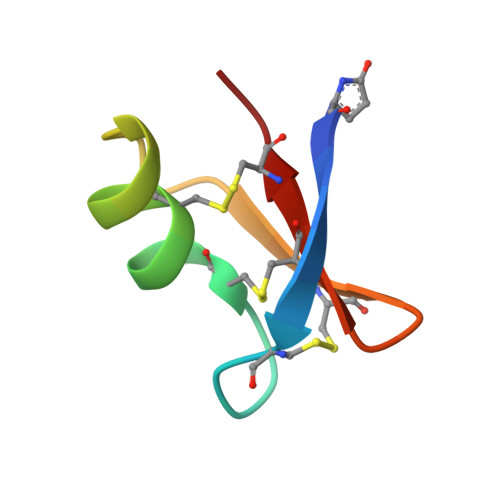
Synthesis by native chemical ligation and characterization of the scorpion toxin AmmTx3.
Zoukimian, C., Meudal, H., De Waard, S., Ouares, K.A., Nicolas, S., Canepari, M., Beroud, R., Landon, C., De Waard, M., Boturyn, D.(2019) Bioorg Med Chem 27: 247-253
- PubMed: 30529150
- DOI: https://doi.org/10.1016/j.bmc.2018.12.009
- Primary Citation of Related Structures:
6GGZ - PubMed Abstract:
The scorpion toxin AmmTx3 is a specific blocker of K v 4 channels. It was shown to have interesting potential for neurological disorders. In this study, we report the first chemical synthesis of AmmTx3 by using the native chemical ligation strategy and validate its biological activity. We determined its 3D structure by nuclear magnetic resonance spectroscopy, and pointed out that AmmTx3 possesses the well-known CSαβ structural motif, which is found in a large number of scorpion toxins. Overall, this study establishes an easy synthetic access to biologically active AmmTx3 toxin.
Organizational Affiliation:
Department of Molecular Chemistry, Univ. Grenoble Alpes, CNRS, 570 rue de la chimie, CS 40700, Grenoble 38000, France; Smartox Biotechnology, 6 rue des platanes, Saint Egrève 38120, France.