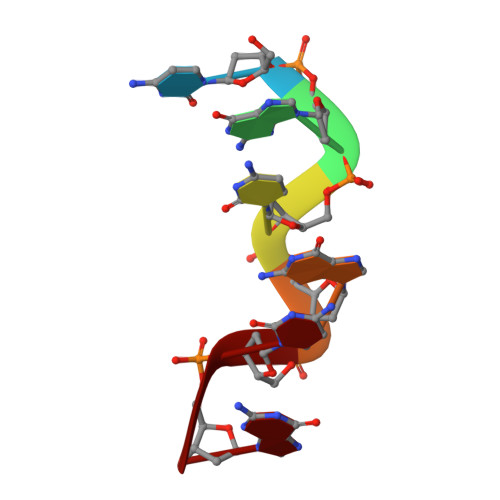
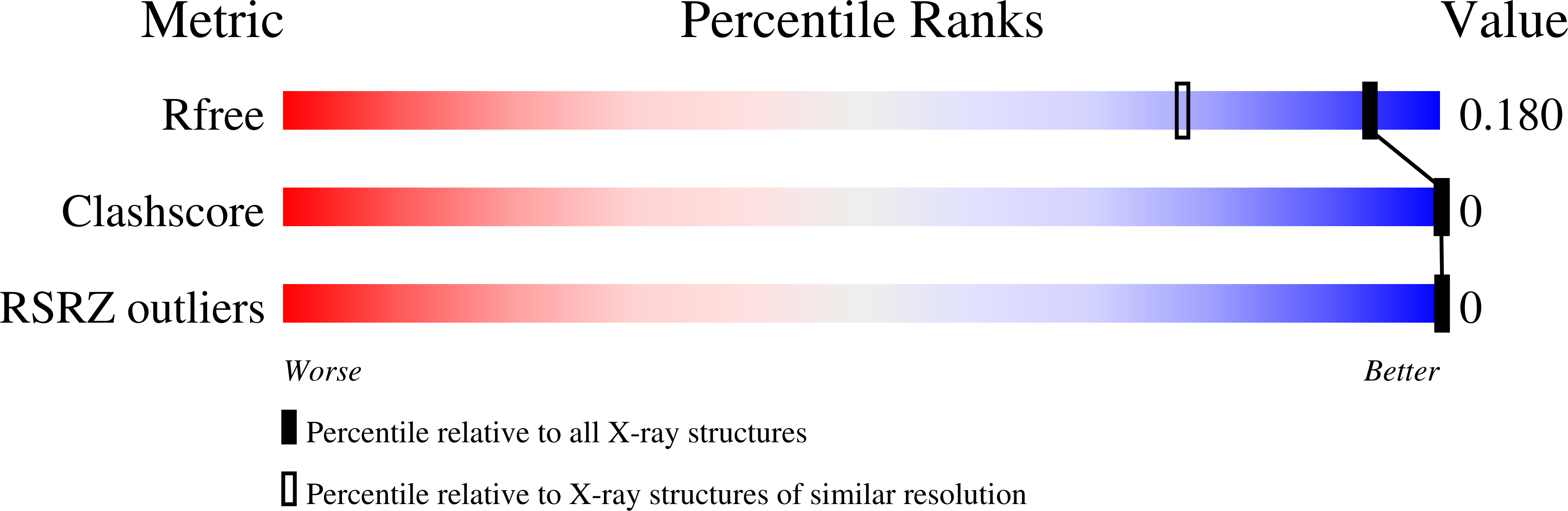The crystal structure of Z-DNA with untypically coordinated Ca
Luo, Z., Dauter, Z.(2018) J Biol Inorg Chem 23: 253-259
- PubMed: 29270817
- DOI: https://doi.org/10.1007/s00775-017-1526-4
- Primary Citation of Related Structures:
6BST - PubMed Abstract:
DNA oligomer duplexes with alternating cytosines and guanines in their sequence tend to form helices of the Z-DNA type, where the sugar and phosphate backbone forms a left-handed helix in a zigzag fashion with a repeat of two successive Watson-Crick pairs of nucleotides. Z-DNA duplexes often crystallize in complexes with diverse metal ions interacting with polar DNA atoms in various ways. This work describes the high-resolution crystal structure of a Z-DNA d(CGCGCG) 2 duplex in complex with Ca 2+ ions, unusually coordinated as an approximate pentagonal bipyramid by two neighboring guanines through their O6 and N7 atoms and a water molecule in the equatorial plane and a phosphate oxygen atom and another water molecule in the apical positions.
Organizational Affiliation:
Synchrotron Radiation Research Section, MCL, National Cancer Institute, Argonne National Laboratory, Argonne, IL, 60439, USA.