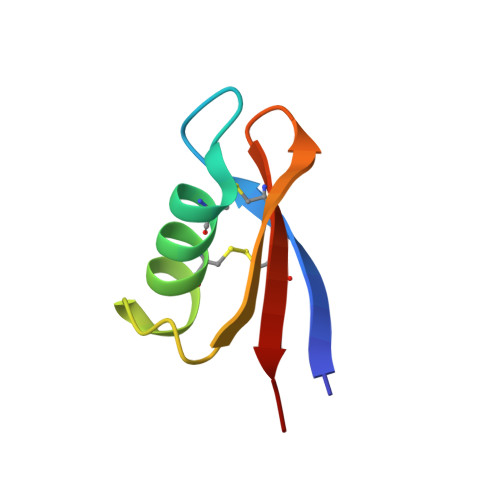
A Centipede Toxin Family Defines an Ancient Class of CS alpha beta Defensins.
Dash, T.S., Shafee, T., Harvey, P.J., Zhang, C., Peigneur, S., Deuis, J.R., Vetter, I., Tytgat, J., Anderson, M.A., Craik, D.J., Durek, T., Undheim, E.A.B.(2019) Structure 27: 315-326.e7
- PubMed: 30554841
- DOI: https://doi.org/10.1016/j.str.2018.10.022
- Primary Citation of Related Structures:
6BL9 - PubMed Abstract:
Disulfide-rich peptides (DRPs) play diverse physiological roles and have emerged as attractive sources of pharmacological tools and drug leads. Here we describe the 3D structure of a centipede venom peptide, U-SLPTX 15 -Sm2a, whose family defines a unique class of one of the most widespread DRP folds known, the cystine-stabilized α/β fold (CSαβ). This class, which we have named the two-disulfide CSαβ fold (2ds-CSαβ), contains only two internal disulfide bonds as opposed to at least three in all other confirmed CSαβ peptides, and constitutes one of the major neurotoxic peptide families in centipede venoms. We show the 2ds-CSαβ is widely distributed outside centipedes and is likely an ancient fold predating the split between prokaryotes and eukaryotes. Our results provide insights into the ancient evolutionary history of a widespread DRP fold and highlight the usefulness of 3D structures as evolutionary tools.
Organizational Affiliation:
Institute for Molecular Bioscience, The University of Queensland, St Lucia, QLD 4072, Australia.