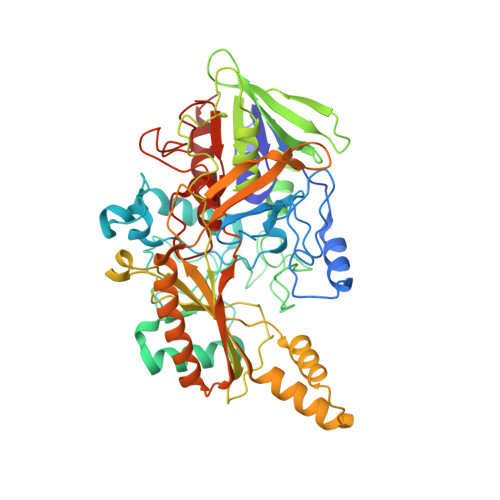
X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.
Yoshida, H., Kojima, K., Shiota, M., Yoshimatsu, K., Yamazaki, T., Ferri, S., Tsugawa, W., Kamitori, S., Sode, K.(2019) Acta Crystallogr D Struct Biol 75: 841-851
- PubMed: 31478907
- DOI: https://doi.org/10.1107/S2059798319010878
- Primary Citation of Related Structures:
6A2U - PubMed Abstract:
The bacterial flavin adenine dinucleotide (FAD)-dependent glucose dehydrogenase complex derived from Burkholderia cepacia (BcGDH) is a representative molecule of direct electron transfer-type FAD-dependent dehydrogenase complexes. In this study, the X-ray structure of BcGDHγα, the catalytic subunit (α-subunit) of BcGDH complexed with a hitchhiker protein (γ-subunit), was determined. The most prominent feature of this enzyme is the presence of the 3Fe-4S cluster, which is located at the surface of the catalytic subunit and functions in intramolecular and intermolecular electron transfer from FAD to the electron-transfer subunit. The structure of the complex revealed that these two molecules are connected through disulfide bonds and hydrophobic interactions, and that the formation of disulfide bonds is required to stabilize the catalytic subunit. The structure of the complex revealed the putative position of the electron-transfer subunit. A comparison of the structures of BcGDHγα and membrane-bound fumarate reductases suggested that the whole BcGDH complex, which also includes the membrane-bound β-subunit containing three heme c moieties, may form a similar overall structure to fumarate reductases, thus accomplishing effective electron transfer.
Organizational Affiliation:
Life Science Research Center and Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan.